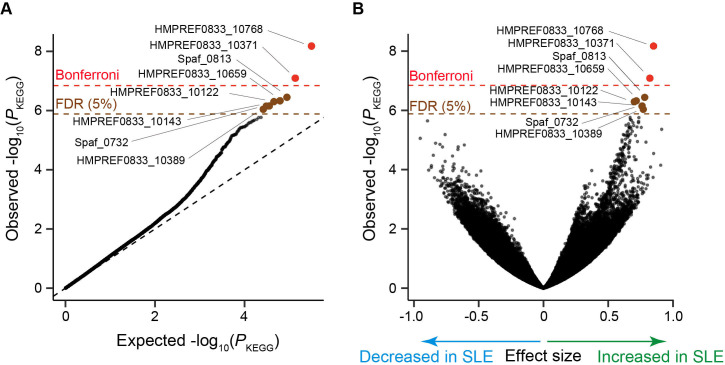
Figure 2.
Result of the SLE MWAS based on the microbial gene abundance data. (A) A quantile-quantile plot of the MWAS p values of the genes (P KEGG). The x-axis indicates log-transformed empirically estimated median P KEGG. The y-axis indicates observed −log10(P KEGG). The diagonal dashed line represents y=x, which corresponds to the null hypothesis. The horizontal red line indicates the empirical Bonferroni-corrected threshold (α=0.05), and the brown line indicates the empirically estimated FDR threshold (FDR=0.05). Genes with P KEGG less than the Bonferroni thresholds are plotted as red dots. Genes with FDR<0.05 are plotted as brown dots, and other clades are plotted as black dots. (B) A volcano plot. The x-axis indicates effect sizes in linear regression. The y-axis, horizontal lines and dot colours are the same as in (A). FDR, false discovery ratio; KEGG, Kyoto Encyclopedia of Genes and Genomes; MWAS, metagenome-wide association study; SLE, Systemic lupus erythematosus.