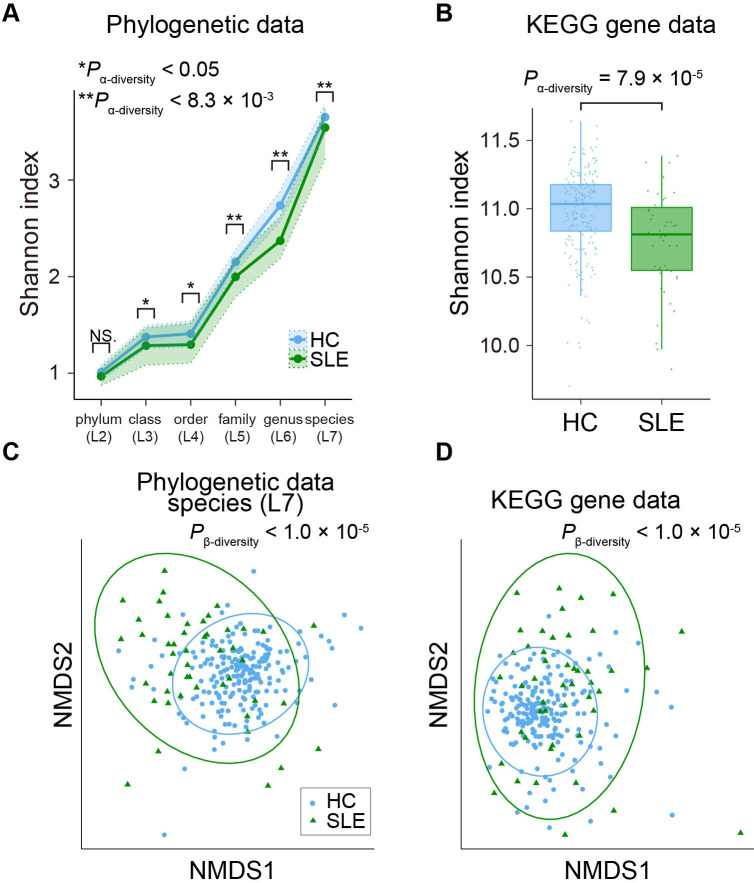
Figure 4.
Case-control comparison of the microbial diversities in SLE. (A) α-diversities of the phylogenetic relative abundance data for the six taxonomic levels. Blue and green dots represent the median Shannon index of the HC and SLE subjects. Upper and lower dashed lines indicate the first and third quantile of Shannon index for the HC and SLE subjects. (B) α-diversities of the gene abundance based on KEGG gene databases. Boxplots indicate the median values (centre lines) and the IQRs (box edges), with whiskers extending to the most extreme points within the range between (lower quantile − [1.5×IQR]) and (upper quantile + [1.5×IQR]). (C) β-diversities of the phylogenetic relative abundance data at the species level. Result of NMDS based on Bray-Curtis distance is represented. Blue and green dots represent the HC and SLE subjects. (D) β-diversities of the gene abundance based on KEGG gene database. Result of NMDS based on Bray-Curtis distance is represented. Blue and green dots represent the HC and SLE subjects. *P α-diversity<0.05; **P α-diversity<0.0083. HC, healthy control; KEGG, Kyoto Encyclopedia of Genes and Genomes; NMDS, non-metric multidimensional scaling; SLE, systemic lupus erythematosus.