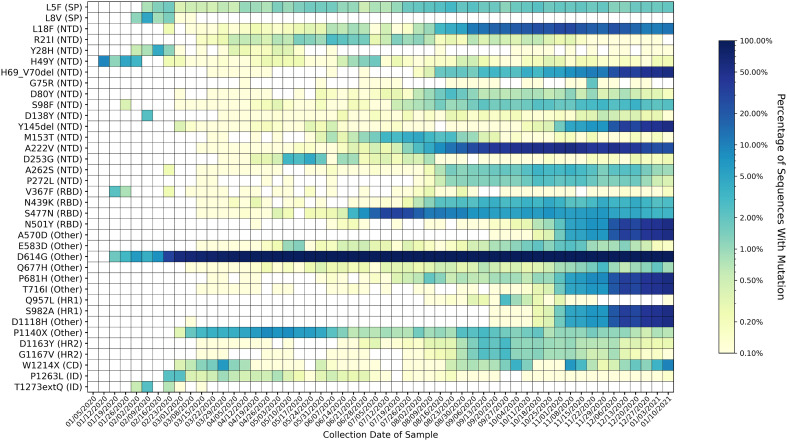
Fig. 2.
Heatmap of all mutations observed in 2% or more of genomes collected for at least one week. Variants are listed on the y-axis and sorted according to their position in the spike protein sequence. Parentheses give the domain in which each variant appears, according to the domain positions specified in Huang et al. 2020 (Graham et al., n.d.). Key for domain abbreviations: SP = signal peptide, NTD=N-terminal domain, RBD = receptor-binding domain, FP = fusion peptide, HR1 = heptad repeat 1, HR2 = heptad repeat 2, CD = cytoplasmic domain, ID = intracellular domain. The heatmap is colored based on a log-10 scale, with prevalence values of zero colored in white, and values less than or equal to 0.10% colored with the lightest shade. Time on the x-axis is categorized by week of collection date, beginning on January 5th, 2020, and ending on January 16th, 2021. Mutations associated with the Alpha variant of concern (H69_V70del, Y145del, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H) have rapidly increased in prevalence since the appearance of the variant in mid-September 2020. Varying trends are observed for other mutations; some are consistently present, while others have emerged and later disappeared. Fifteen out of the 36 variants observed in at least 2% of samples were in the N-terminal domain, four out of 33 were in the receptor-binding domain, two each were in the signal peptide, the intracellular domain, and heptad repeat 2; one was in the cytoplasmic domain, and eight were in an unspecified domain (other).