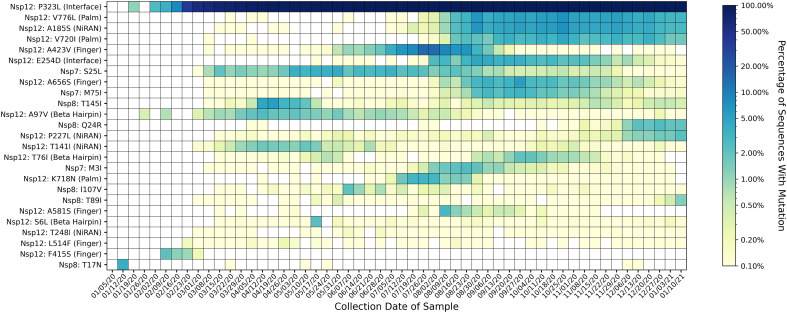
Fig. 7.
Heatmap of all mutations observed in 2% or more of genomes collected for at least one week. Mutations are listed on the y-axis, and parentheses give the domain in which each mutation appears, according to the domain ranges specified in Yin et al. 2020 (Madhi et al., 2021). The heatmap is colored based on a log-10 scale, with prevalence values of zero colored in white, and values less than or equal to 0.10% colored with the lightest shade. Time on the x-axis is categorized by week of collection date, beginning on January 5th, 2020, and ending on January 16th, 2021. Most mutations in the polymerase complex do not appear to persist, except for the Nsp12 substitutions P323L, V776L, A185S, and V720I. Of the 25 mutations observed in at least 2% of sequences in any week, 17 are in Nsp12, five are in Nsp8, and three are in Nsp7. Of the 17 Nsp12 mutations, five are in the finger subdomain, four are in the NiRAN domain; three each are in the palm subdomain and the Beta Hairpin; two are in the interface domain, and zero are in the thumb subdomain.