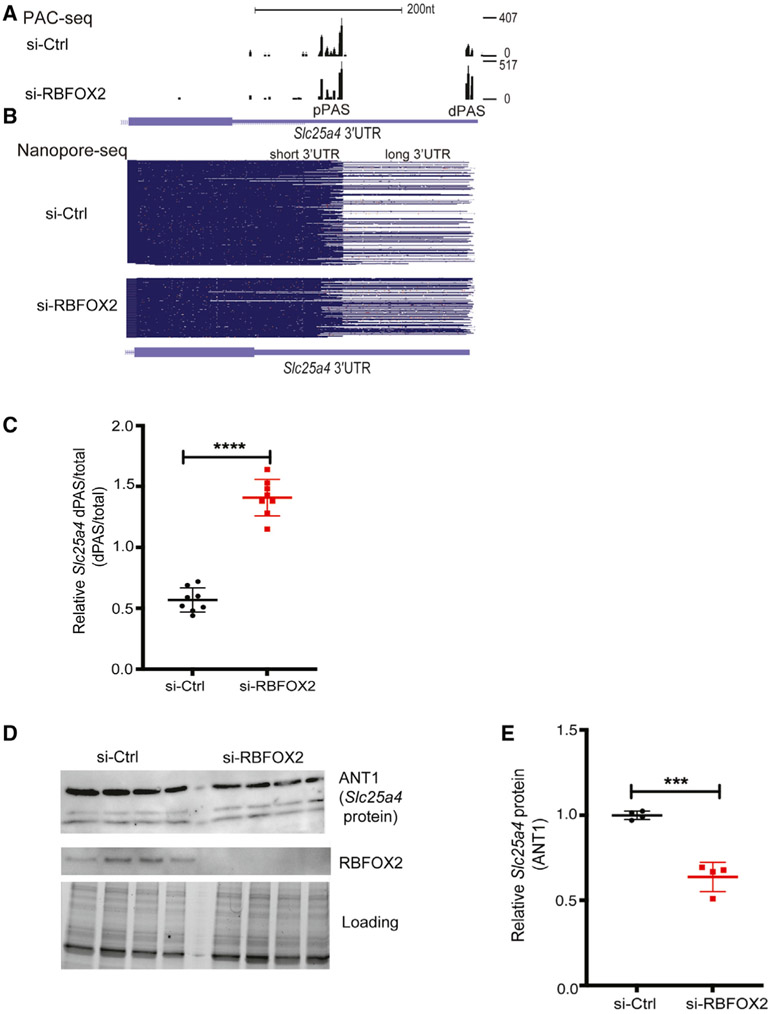
Figure 4. RBFOX2 regulates tandem APA and expression of essential mitochondrial gene Slc25a4.
(A) Slc25a4 3′UTR lengthening mediated via tandem APA in RBFOX2-depleted H9c2 myoblasts identified by PAC-seq. The number of reads mapped to Slc25a4 transcripts is shown on the right side. See also Figure S5.
(B) Nanopore sequencing analysis of full-length Slc25a4 transcripts with different 3′UTR lengths in control versus RBFOX2 KD myoblasts. See also Figure S5.
(C) RT-PCR analysis of endogenous Slc25a4 dPAS/total mRNA ratio in RBFOX2 KD myoblasts ectopically expressing GFP or GFP-RBFOX2. n = 8, ****p < 0.0001 (three independent experiments). See Table S2 for primer information.
(D) WB analysis of ANT1 protein (Slc25a4) in control versus RBFOX2 KD myoblasts determined by Bio-Rad ChemiDoc Imager. Even protein loading was monitored by imaging stain-free gels. ANT1 protein levels were normalized to loading control, and fold change in ANT1 protein levels was quantified using Bio-Rad ChemiDoc software. n = 4, ***p = 0.0002 (three independent experiments).