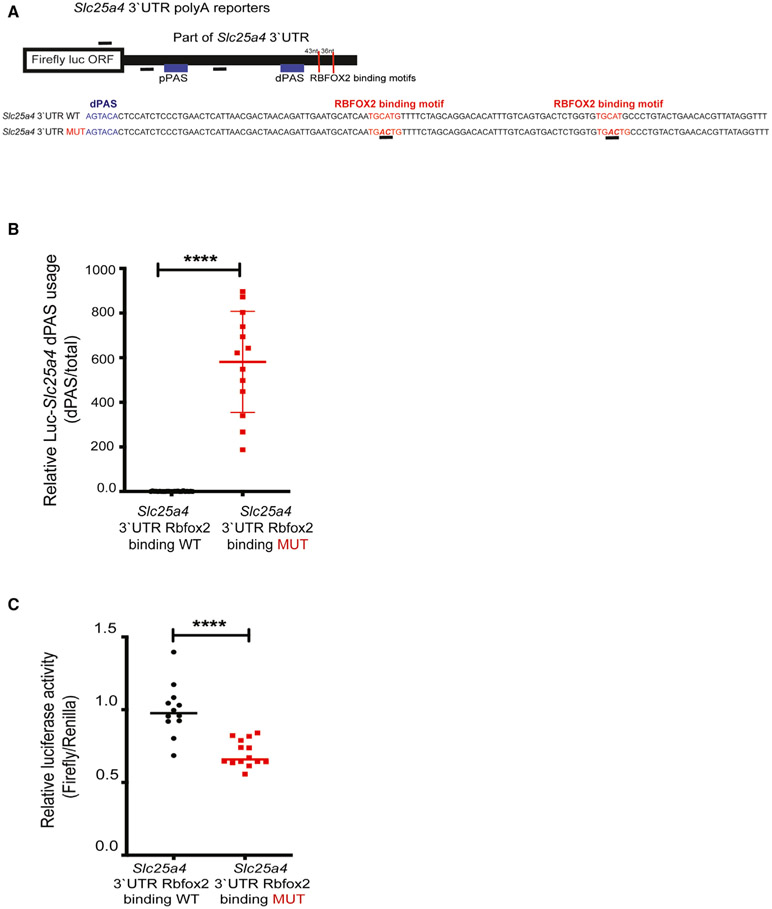
Figure 5. RBFOX2 binding sites near dPASs are critical for APA regulation of the Slc25a4 gene.
(A) Cartoon representation of luciferase-Slc25a4 poly(A) reporter constructs that harbor both pPASs and dPASs of Slc25a4 in the 3′UTR with or without mutated RBFOX2 binding motifs (underlined) located downstream of the dPAS.
(B) qRT-PCR analysis of dPAS usage when normalized to total mRNA levels in HEK293 cells expressing WT or RBFOX2 binding site mutant poly(A) luciferase-Slc25a4 3′UTR constructs. n = 24 for WT, n = 13 for mutant samples from three independent experiments, ****p < 0.0001.
(C) Relative firefly luciferase levels in HEK293 cells expressing WT or RBFOX2 binding site mutant poly(A) luciferase-Slc25a4 3′UTR constructs. n = 12 for WT samples, n = 14 for mutant samples from three independent experiments, ****p < 0.0001.