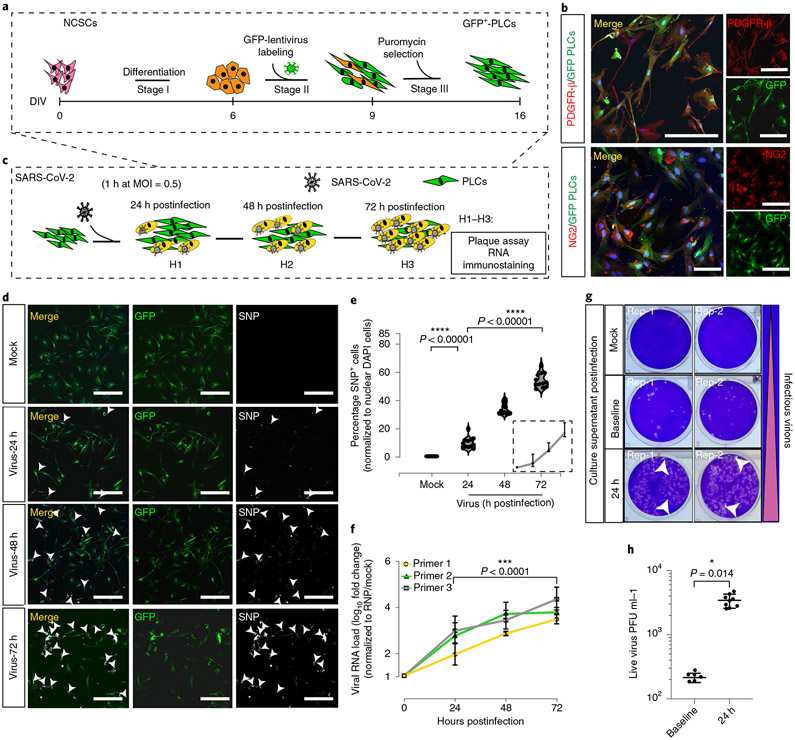
Fig. 1 ∣. SARS-CoV-2 productively infects PLCs.
a, GFP+ PLC generation protocol. b, Immunostaining of GFP+ PLCs shows expression of the pericyte markers PDGFR-β and NG2. GFP+ PLCs cultured on coverslips were fixed and immunostained (DAPI, blue). Scale bar, 100 μm. n = 12 includes 3 biological replicates (3 independent PLC cultures) and 4 technical replicates (4 different image regions). c, SARS-CoV-2 infection of PLC protocol. At each time point, RNA was isolated; in parallel, cells were fixed and immunostained. d, SARS-CoV-2-nucleocapsid protein covisualized with GFP. Nuclei stained with DAPI. Scale bar, 400 μm. GFP, PLCs. Arrowheads, SARS-CoV-2 nucleocapsid protein+ cells. n = 12 includes 3 biological replicates (3 independent PLC cultures) and 4 technical replicates (4 different image regions). e, Quantification of SARS-CoV-2 nucleocapsid protein expression from d. SARS-CoV-2 nucleocapsid protein+ cells were counted in ImageJ. Nuclear DAPI was used as reference. n = 12 includes 3 PLC coverslips from 3 individual biological cultures and 4 image regions from each coverslip. Two-tailed t-test with Šidák multiple comparisons correction was used to determine significance. ****p < 0.00001, P = 0.00000684 for 24 h versus mock and P = 0.00000193 for 72 h versus 24 h postinfection. f. RT-qPCR for SARS-CoV-2 shows increasing viral RNA load over time. RNP was used as reference. n = 12 includes PLCs from 3 individual cultures and 4 technical replicates for each biological replicate. Two-tailed t-test with Šidák multiple comparisons test correction was used to calculate significance. ***P < 0.0001, P = 0.0000749. g, Plaque assay showing increased infectious virions in culture supernatant 24 h postinfection compared to baseline. Mock was used as negative control. Two replicates (rep-1, rep-2) from each condition are shown. h, Plaque assay indicating secretion of live virus into culture supernatant at 24 h postinfection; n = 9 includes 3 replicates generated from 3 different biological replicates (3 individual PLC cultures) and 3 technical replicates for 24 h; n = 6 for baseline infection includes 2 biological replicates (2 different cultures) and 3 technical replicates, P = 0.014, *P < 0.1. Two-way ANOVA with Šidák multiple comparisons test was used for statistics. PLCs were derived from NCSCs generated with H9 cells. e,f,h, The error bars represent the mean ± 1 s.d.