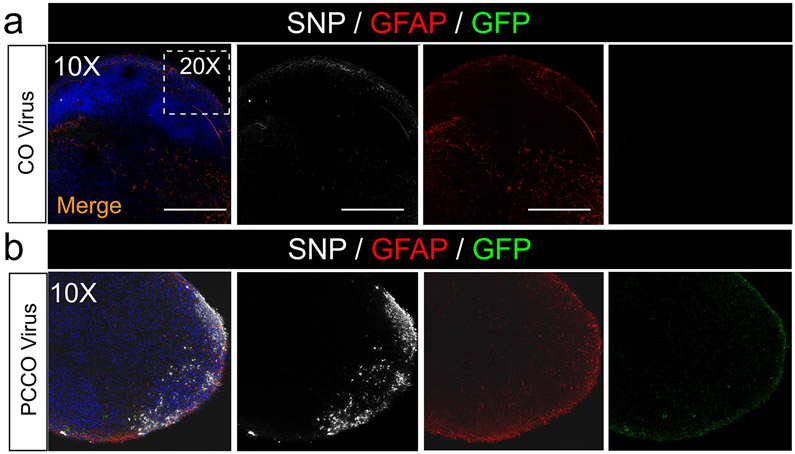
Extended Data Fig. 8 ∣. PCCOs show robust SARS-CoV-2 infection compared to COs.
a-b, SNP and GFAP staining shows few SNP+ cells in COs (a) and robust SNP+ cells in PCCOs (b) 72 h after SARS-CoV-2 virus exposure. Bar: 400 μm. DAPI: blue, GFP: PLCs, GFAP: astrocytes, and SNP: SARS-CoV-2-NP. The dashed white boxes show the 20x region shown in Fig. 3b-e. Bar: 400 μm. In a-b, n = 9 includes three biological replicates (COs generated from H1, iPSC-Line2, iPSC-Line3) and three technical replicates (three COs for each of the biological replicates). Both COs and PCCOs shown were generated from H1 cells and GFP-PLCs were derived from H9 cells.