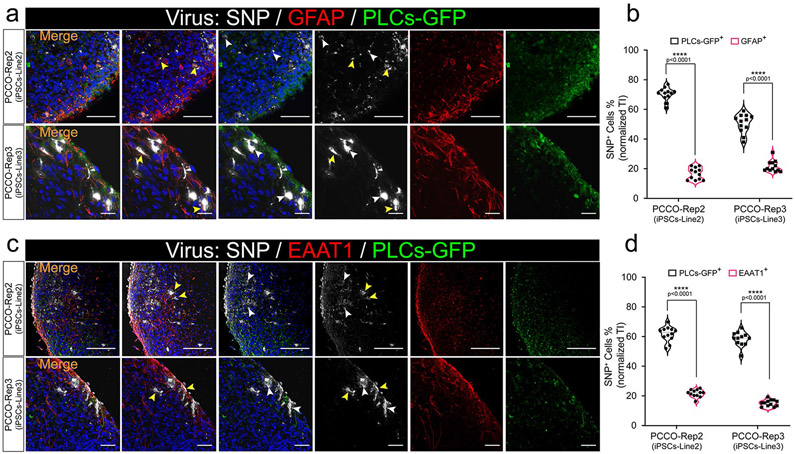
Extended Data Fig. 10 ∣. Both astrocytes and PLCs show evidence of SARS-CoV-2 infection in PCCO.
a, SNP and GFAP staining shows infection of both PLCs and astrocytes within PCCOs derived from different hiPSC lines (iPSC-Line2 and iPSC-Line3) 72 h after SARS-CoV-2 virus exposure. GFAP in red, SNP in gray, yellow arrows: GFAP+/SNP+ cells, white arrows: GFP+/SNP+ cells, bar=400 μm, DAPI in blue. b, Quantification of SNP+/GFAP+ and SNP+/GFP+ cells shown in a. iPSC-Line2 and iPSC-Line3 are shown respectively. n = 12 includes 3 independent biological replicates from different COs/PCCOs and 4 different image regions from at least 3 different section regions in b. Two tailed t-test was used to determine the significance followed by a Sidak multiple-comparison test correction. **** p < 0.0001, p = 0.000067 for PCCO-Rep2 (iPSC-Line2) and p = 0.000097 for PCCO-Rep3 (iPSC-Line3). c, SNP and EAAT1 staining shows infection of both PLCs and astrocytes within PCCOs derived from different hiPSC lines (iPSC-Line2 and iPSC-Line3) 72 h after SARS-CoV-2 virus exposure, EAAT1 in red, SNP in gray, yellow arrows: EAAT1+/SNP+ cells, white arrows: GFP+/SNP+ cells, bar=400 μm, DAPI in blue. d, Quantification of SNP+/EAAT1+ and SNP+/GFP+ cells shown in c. iPSC-Line2 and iPSC-Line3 n = 12 includes 3 independent biological replicates from different COs/PCCOs and 4 different image regions from at least 3 different section regions in b. Two tailed t-test was used to determine the significance followed by a Sidak multiple-comparison test correction. **** p < 0.0001, p = 0.000047 for PCCO-Rep2 (iPSC-Line2) and p = 0.00027 for PCCO-Rep3 (iPSC-Line3). In this figure, PCCOs are derived from different iPSC lines accordingly (see detailed information in Extended Data Fig. 3), and GFP-PLCs are generated from H9 cells.