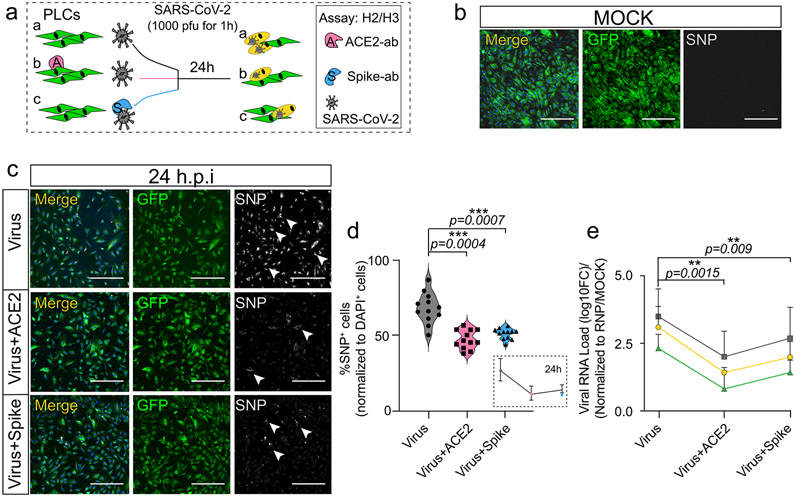
Extended Data Fig. 2 ∣. ACE2 antibody inhibits SARS-CoV-2 infection of PLCs.
a, Schematic of ACE2 and SARS-CoV-2 spike antibody blocking SARS-CoV-2 infection of PLCs protocol. PLCs were pre-incubated with 20μg/ml ACE2 antibody or lgG negative control 30 min before infection with 1000 pfu SARS-CoV-2. As a positive control, virus was pre-incubated with anti-SARS-CoV-2 spike antibody for 30 min at 37°C before infection of PLCs. and 100μg/ml SARS-CoV-2 spike protein antibody for 1 h before infection. RNA was isolated and in parallel cells were fixed and immunostained at 24 hours post infection (h.p.i). (H2: qPCR, H3: immunostaining). b-c, SARS-CoV-2-Nucleocapsid protein (SNP) co-visualized with GFP. DAPI: nuclei, Bar: 400 μm. GFP: PLCs, Arrows: SNP+ cells. MOCK infection shown in b. SARS-CoV-2 infection (virus), SARS-CoV-2 blocking with ACE2 antibody, and SARS-CoV-2 blocking with SARS-CoV-2 Spike antibody shown in c respectively. n = 12 includes three biological replicates (three independent PLCs cultures) and four technical replicates (Four different image regions). d, Quantification of SNP expression from c. SNP+ cells were counted in ImageJ, nDAPI used as reference. n = 12 includes 3 PLC cover slips from three different biological PLCs cultures and 4 image regions from each cover slip. Two tailed t-test with Sidak multiple-comparison correction was used to determine the significance. *** p < 0.001. e, qRT-PCR for SARS-CoV-2 shows reduction viral RNA load following pre-incubation with ACE2 and spike blocking antibodies. Gray, green, and yellow represent three separate CDC-designated primer sets used for RT-qPCR. RNP used as reference. n = 12 includes PLCs from 3 different wells and 4 technical replicates for each biological replicate. Two tailed t-test with Sidak multiple-comparison test correction was used to calculate significance ** p < 0.01, error bar represents mean±1 SD.