Fig. 1.
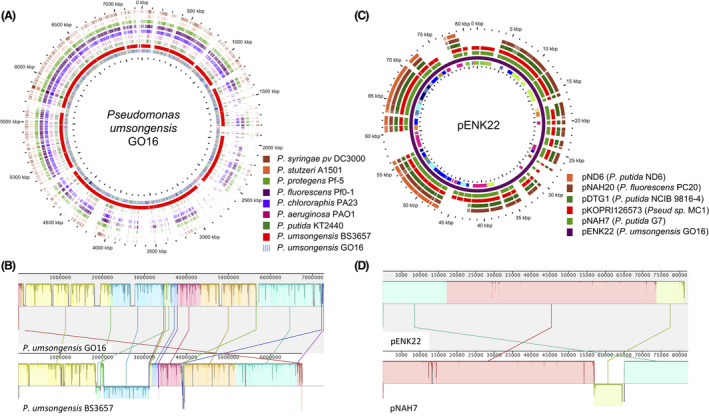
Global analysis of the P. umsongensis GO16 genome.
A. BLAST atlas comparing genes present in strain GO16 (used as a reference) with other representative Pseudomonas strains. The circles represent the genomes of the species shown in the insert starting with Pseudomonas syringae pv tomato DC3000 (outer circle) and finishing with P. umsongensis GO16 shown as the set of clusters of orthologous groups (COGs; inner circle).
B. Genome alignment of strains of P. umsongensis GO16 (upper bar) and BS3657 (lower bar).
C. BLAST atlas comparing present genes in plasmid pENK22 with other representative plasmids of Pseudomonas species containing naphthalene‐catabolic genes (nah).
D. Detailed alignment between pENK22 and the naphthalene degrading plasmid NAH7. Genes, COG (represented by the innermost circle) and blast analysis resulting from the analysis are summarized in Tables S1 and S2.
