Fig. 1.
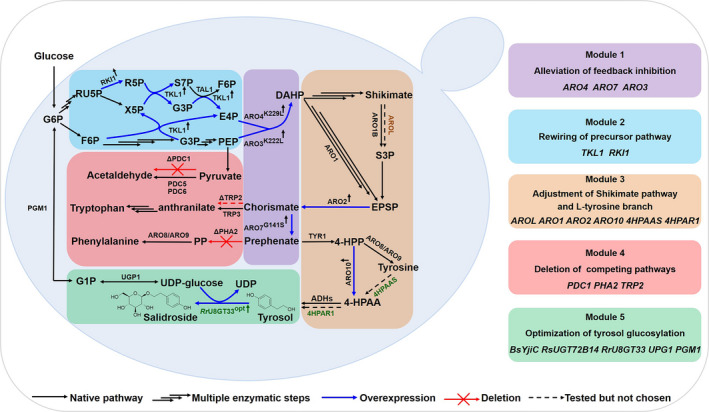
Schematic representation of the modular engineering strategy for high‐level tyrosol and salidroside production in S. cerevisiae. The modular engineering strategy consists of alleviation of feedback inhibition (module 1), rewiring of precursor pathway (module 2), adjustment of shikimate pathway and l‐tyrosine branch (module 3), deletion of competing pathways (module 4) and optimization of tyrosol glycosylation (module 5). Black arrows represent the native pathways in S. cerevisiae; blue bold arrows represent the overexpressed genes in this study; red crosses represent gene deletions; dashed arrows indicate that the reactions were tested but not used in this study and the treble arrows represent multiple enzymatic steps. Native genes of S. cerevisiae are shown in black colour; the gene AROL from E. coil is indicated in orange; the heterologous genes from Rhodiola rosea were shown in green. Metabolite abbreviations: G6P: Glucose 6‐phosphate; RU5P: D‐Ribulose 5‐phosphate; X5P: D‐Xylulose 5‐phosphate; R5P: D‐Ribose 5‐phosphate; S7P: Sedoheptulose 7‐phosphate; G3P: D‐Glyceraldehyde 3‐phosphate; F6P: beta‐D‐Fructose 6‐phosphate; PEP: phosphoenolpyruvate; E4P: D‐Erythrose 4‐phosphate; DAHP: 3‐deoxy‐D‐arabino‐heptulosonate‐7‐phosphate; S3P: shikimate‐3‐phosphate; EPSP: 5‐enolpyruvyl‐3‐shikimate; 4‐HPP: p‐hydroxyphenylpyruvate; PP: phenylpyruvate; 4‐HPAA: 4‐hydroxyphenylacetylaldehyde; G1P: Glucose 1‐phosphate; UDP: Uridine diphosphate; Gene abbreviations: RKI1: ribose‐5‐phosphate ketol‐isomerase1; TKL1: transketolase 1; TAL1: transaldolase 1; ARO3K222L/ARO4K229L : feedback‐insensitive DAHP synthases; ARO1: pentafunctional aromatic enzyme; AROL: shikimate kinase Ⅱ from E. coil; ARO2: chorismate synthase; ARO7G141S : feedback‐insensitive chorismate mutase; PHA2: prephenate dehydratase; TRP2: anthranilate synthase; TRP3: bifunctional anthranilate synthase/indole‐3‐glycerol‐phosphate synthase; PDC1: pyruvate decarboxylase 1; PDC5: pyruvate decarboxylase 5; PDC6: pyruvate decarboxylase 6; TYR1: prephenate dehydrogenase; ARO10: phenylpyruvate decarboxylase; ARO8/9: aromatic amino transferases; ADHs: alcohol dehydrogenases; 4HPAAS: 4‐HPAA synthase from Rhodiola. rosea; 4HPAR1: 4‐HPAA reductase 1 from Rhodiola. rosea; RrU8GT33: tyrosol: UDP‐glucose 8‐O‐glucosyltransferase 33 from Rhodiola. rosea; PGM1: Phosphoglycerate mutase 1; UPG1: UDP‐glucose pyrophosphorylase 1.
