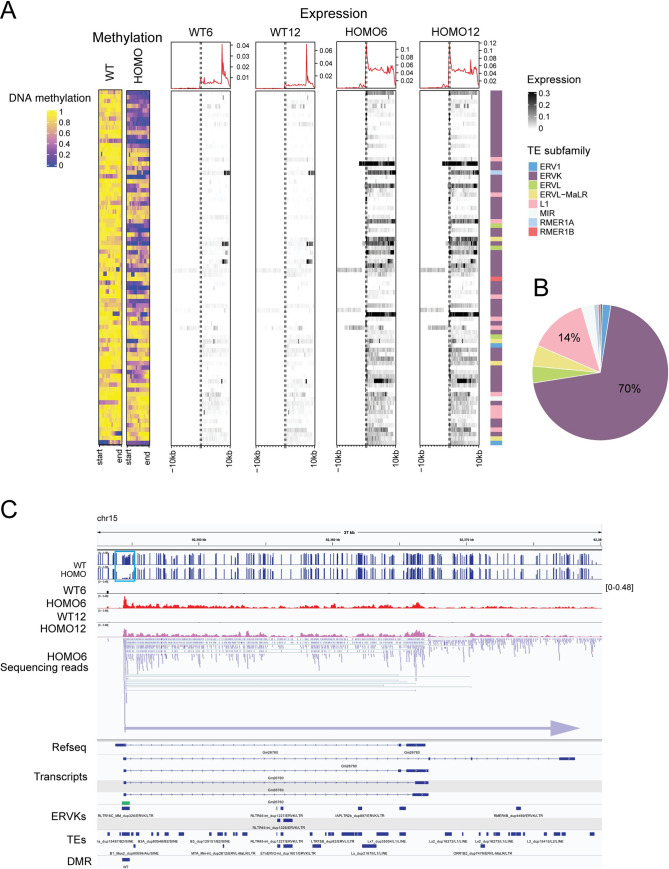
Fig 9. EHMT2 is required for DNA methylation targeting and suppression of transcription from transposable elements predominantly of the ERVK-class.
(A) Heatmaps of differentially methylated TEs associated with differentially expressed long transcripts. DNA methylation is displayed at TE-DMRs in WT and HOMO embryos (to the left) and transcription is displayed along 10 kilobases before and after each TE in the embryos marked at the top. Normalized expression levels (logCPM) and the TE families are shown to the right. (B) Pie chart displays the proportion of DE repeat subfamilies associated with DNA methylation change using the color code as above. (C) Example for transcription initiation from an ERVK DMR in HOMO6 and HOMO12 embryos. IGV browser images of WGBS and RNAseq experiments are displayed in samples indicated to the left. The coverage is displayed for one WT6, HOMO6, WT12 and HOMO12 embryo, and the sequencing reads are displayed for one HOMO6 sample. Tracks of Refseq transcripts, putative transcripts, ERVKs, TEs, and DMRs are also included as marked to the left. A blue horizontal arrow indicates the long noncoding transcript in chr15 defined by sequencing reads (all blue) in the sense direction. It starts in an RLTR16C ERVK repeat (green rectangle), which is a DMR (turquoise rectangle), and it matches the putative transcript Gm26760. It encompasses multiple ‘passenger’ repeats, which also appear to be DE repeats, but which only display sequencing reads in the sense direction (blue) irrespective of their initial directionality.