Figure 3.
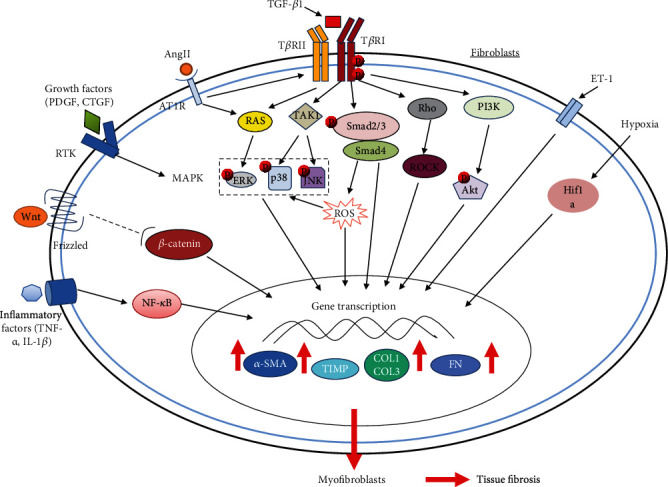
Molecular mechanisms in tissue fibrosis. The diagram shows the TGF-β1, AngII, ET-1, growth factors (PDGF, CTGF, etc.), inflammatory factors (TNF-α, IL-1β, etc.), Wnt, ROS, and hypoxia-inducible factor-1α (HIF-1α) pathways that may mediate tissue fibrotic responses. The central pathways for tissue fibrosis are TGF-β1 canonical (Smad-dependent) and noncanonical (Smad-independent) signaling pathways, among which the canonical TGF-β1/Smad pathway plays a major role in the development of fibrosis. Following TGF-β1 binding, type II TGF-β1 receptor (TβRII) recruits type I TGF-β1 receptor (TβRI) and activates it by phosphorylating it. The activated TβRI then specifically phosphorylates Smad2 and Smad3, which then bind to Smad4 to form a complex leading to their translocation to the nucleus and regulation of transcription of profibrotic genes. Apart from Smad-mediated signal transduction, TGF-β1 can also signal through several noncanonical signaling cascades such as PI3K, p38, ERK, JNK, and Rho-like GTPase pathways. Most of the other pathways have been indicated to regulate or to interact with the TGF-β1 signaling pathways. The final result of these signaling pathways activation is triggering a profibrotic gene transcriptional regulation program contributing to tissue fibrosis caused by the activation of myofibroblasts and their increased synthesis of various myofibroblast-specific and profibrotic proteins such as α-SMA, COL1, COL3, FN, and tissue inhibitors of metalloproteinase (TIMP) [Adapted from ref. [15]].
