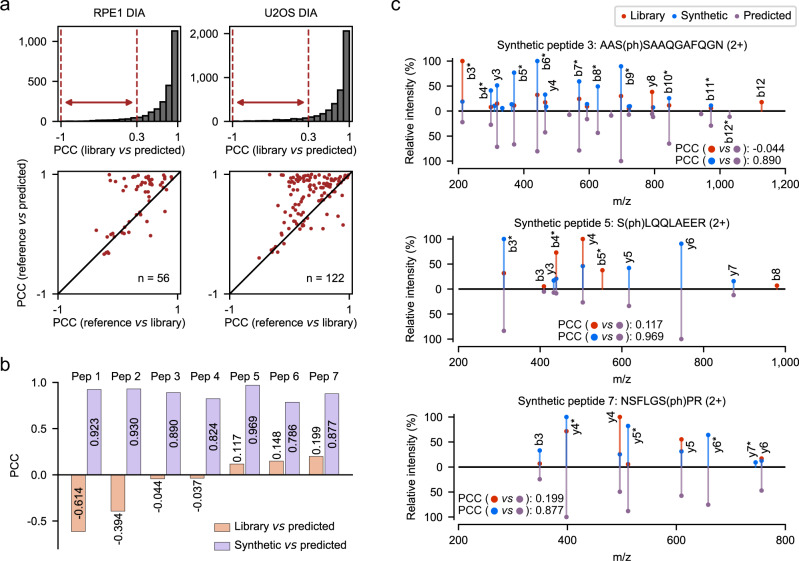
Fig. 2. MSMS spectra prediction by DeepPhospho pinpoints possibly false identifications in an experimental library.
a Distribution of PCC between spectra predicted by DeepPhospho vs spectra assigned in the DIA library for two datasets (upper). For phosphopeptides of low spectral similarity (PCC within −1 to 0.3), their PCC distribution are calculated between predicted spectra vs reference spectra, and between predicted spectra vs DIA library spectra, and plotted around the diagonal (lower). Reference spectra were obtained by gold-standard DDA analysis of the same phosphopeptide samples. b Correlation between the predicted spectra and the high-quality spectra of the synthetic peptide, and between the predicted spectra and the DIA library spectra, for seven selected phosphopeptides. c Spectra mirror plots for phosphopeptides show much higher similarity between the predicted spectra, and the synthetic peptide spectra than between the predicted spectra and the DIA library spectra. Relative fragment ion intensities in the predicted spectra, the DIA library spectra and the synthetic peptide spectra are annotated by purple, orange and blue lines. * Indicates the loss of a phosphate. Source data for this figure are provided as a Source data file.