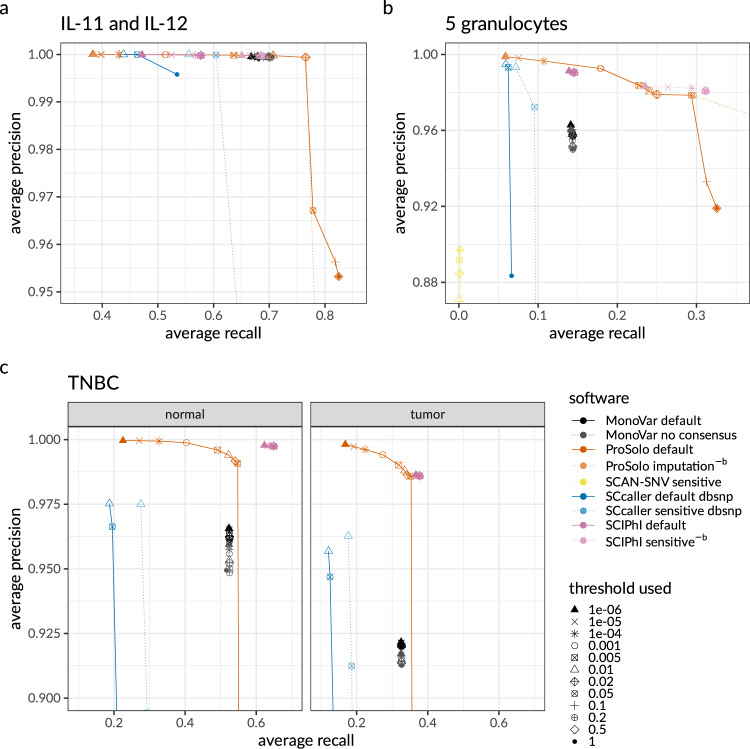
Fig. 1. Precision-recall plots for alternative allele calls of ProSolo, MonoVar, SCAN-SNV, SCcaller, and SCIPhI.
All panels are strong zoom-ins, focusing on (different) areas of interest. Global views of these panels are provided in Supplementary Fig. 7. a Precision and recall of an average of two whole genome sequenced single cells IL-11 and IL-12 against their kindred clone IL-1C as ground truth genotypes. b Precision and recall average of the five whole-exome sequenced single granulocytes against their pedigree-based germline genotype ground truth. c Precision and recall average of 16 tumor and 16 normal single cells sequenced at the whole exome level. −b The germline ground truth induces an artificial increase of recall for SCIPhI’s sensitive and ProSolo’s imputation mode; these modes should thus be disregarded for a fair comparison on the granulocyte dataset in panel b. Threshold parameters (not comparable across tools): MonoVar --t; ProSolo --fdr; SCAN-SNV --fdr; SCcaller -a cutoff; SCIPhI prosolo --fdr. Software modes: MonoVar with consensus filtering (default) or without (no consensus); ProSolo with minimum coverage 1 in single-cell (default), or imputing zero coverage sites based on bulk sample (imputation); SCcaller with recommended settings (default) or with a more sensitive calling; SCIPhI with default parameters (default) or all heuristics off (sensitive).