Fig. 3. High-level overview of ProSolo’s variant calling model.
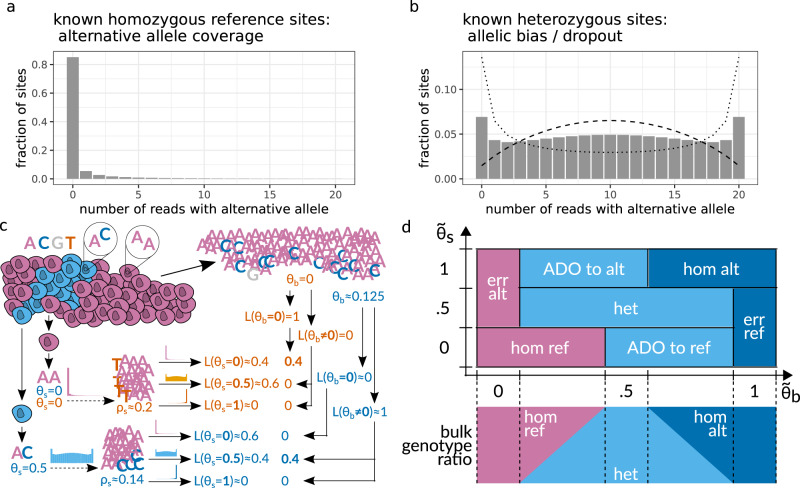
a, b Exemplary alternative allele read count distributions for sites covered by 20 reads, as derived by Lodato et al.22 Homozygous reference sites in a are assumed to follow a beta-binomial distribution; sites heterozygous for the alternative allele in b are assumed to follow the linear combination of two symmetrical beta-binomial distributions (dotted and dashed lines). c Toy example of calling the same genomic site in two single cells from the same population that differ in their true underlying allele frequencies for alternative allele C (blue, θs = 0 vs. θs = 0.5). Alternative nucleotide T (orange) is an amplification error. Empirical distributions in A and B account for the amplification bias, and likelihoods for the alternative allele candidates from the bulk reduce the likelihoods of amplification errors, thereby correctly identifying both the error and the original true mutation. This is formalized with the model in D. d Definition of single-cell events based on ProSolo’s likelihood density estimates for the spectrum of true underlying alternative allele frequencies in the single-cell () and the bulk (). The bulk is always assumed to be a combination of a maximum of two genotypes at a particular site, generating all possible θb (bottom panel). The model further assumes that the bulk sample has sufficient coverage to capture somatic variants. ADO allele dropout, alt alternative, err error, het heterozygous, hom homozygous, ref reference.