Fig. 4. Benchmark Datasets.
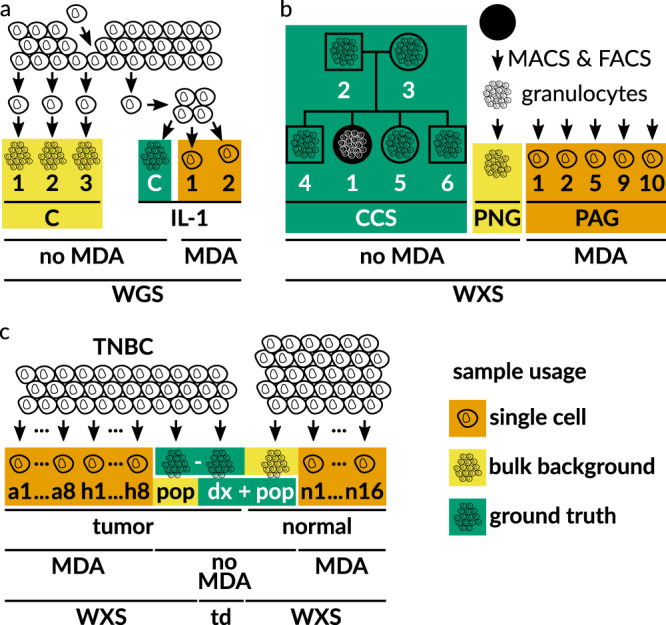
For single cells, DNA was multiple displacement amplified (MDA). a Whole genome sequencing (WGS) dataset, generated from a clonal population started with an individual cell28, and expanded further downstream to generate different bulk samples (C1, C2, C3, and IL-1C). b Newly generated whole exome sequencing (WXS) dataset of blood cells. Ground truth genotypes of patient CCS1 were determined from sequencing data of family members (boxes male; circles female). Granulocytes were isolated from blood using magnetic and fluorescence-activated cell sorting (MACS and FACS). c Whole exome sequencing (WXS) dataset from a triple-negative breast cancer (TNBC) patient21. For tumor cells (a1 to a8 and h1 to h8), the cell population (pop) sequencing of the normal sample was taken as the ground truth, including known clonal tumor-specific somatic variants previously validated by deep targeted duplex (td, dx) sequencing21. For normal cells (n1 to n16), the cell population (pop) sequencing of the tumor sample was taken as the ground truth, after removing known clonal and subclonal tumor-specific somatic variants previously validated by deep targeted duplex (td, dx) sequencing21.
