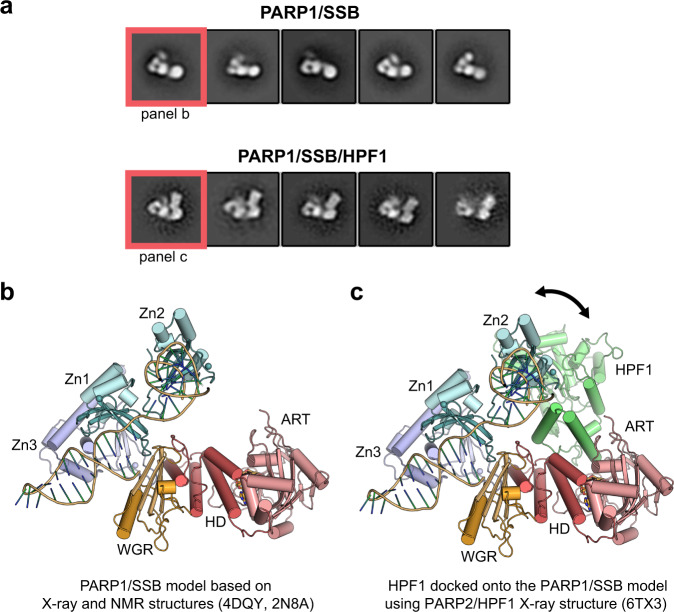
Fig. 7. Architecture of the HPF1 complex with full-length PARP1 bound to a DNA break.
The PARP1/DNA SSB complex was analyzed by negative-stain electron microscopy in the absence and presence of HPF1. a 2D class averages provided views of the complexes without and with HPF1 that could be interpreted with the aid of published structures of PARP1 domains (Zn1, Zn2, Zn3, WGR, and CAT (HD/ART)) on DNA as shown in b and c. b A combined model of a PARP1 Zn1/Zn3/WGR-CAT/DNA structure (4DQY) aligned to a PARP1 Zn1/Zn2/DNA NMR structure (2N8A) c Same as in b with HPF1 added based on the structure of PARP2 CATΔHD/HPF1 (6TX3)43. The 2D averages did not suggest a location for the BRCT domain, which we infer to remain flexibly tethered to the rest of the complex. Consistent with our interpretation of PARP1 domains, the addition of HPF1 adds to the globular end of the protein interpreted as the ART (panel c). The orientation of HPF1 with respect to the catalytic domain remains relatively fixed, whereas the orientation of HPF1 relative to the PARP1 regulatory domains appears to adopt two different conformations, highlighted by the arrow in panel c.