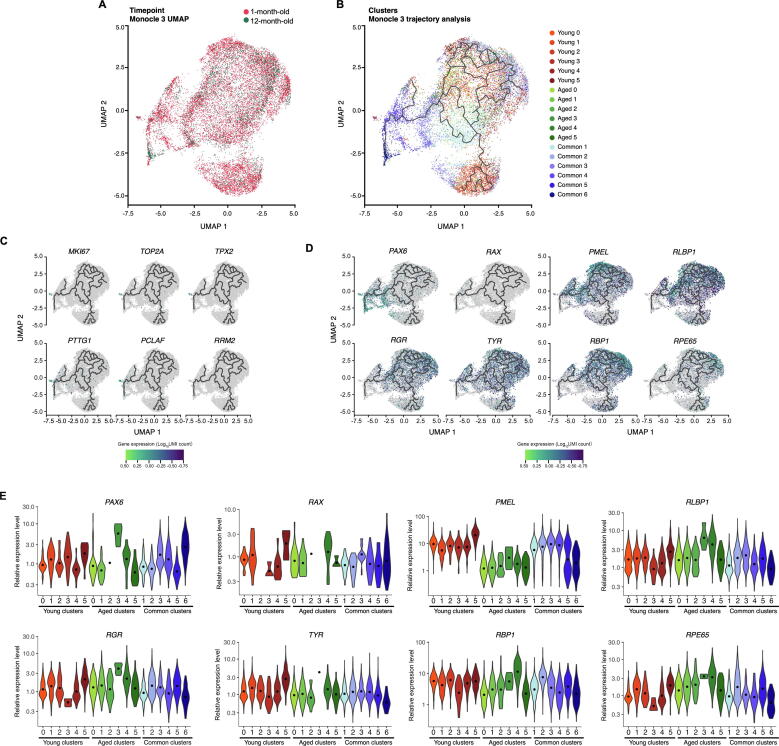
Figure 5.
Only a few cells retain a proliferative profile
Highly dimensional expression data were reduced into two dimensions using UMAP. Plot axes (UMAP1 and UMAP2) represent coordinates in the resulting 2D space. A. UMAP of single cell expression profile split by conditions (1-month-old and 12-month-old). B. UMAP of single cell expression profile for trajectory analysis using Monocle 3. C. UMAP of single cell expression profile for markers associated with proliferation (MKI67, TOP2A, PCLAF, RRM2, TPX2, and PTTG1) across all cells. Expression levels measured as Log10 normalised UMI counts are represented by colour intensity. Lines represent differentiation trajectories as calculated by Monocle3. D. Pseudotime analysis of early retinal markers (PAX6 and RAX) and RPE genes (PMEL, RLBP1, RGR, TYR, RBP1, and RPE65) across the 1-month-old and 12-month-old cells. Expression levels measured as Log10 normalised UMI counts are represented by colour intensity. Lines represent differentiation trajectories as calculated by Monocle3. E. Violin plots of normalised UMI counts of early retinal markers and RPE genes across all subpopulations showing variations in expression across cluster groups.