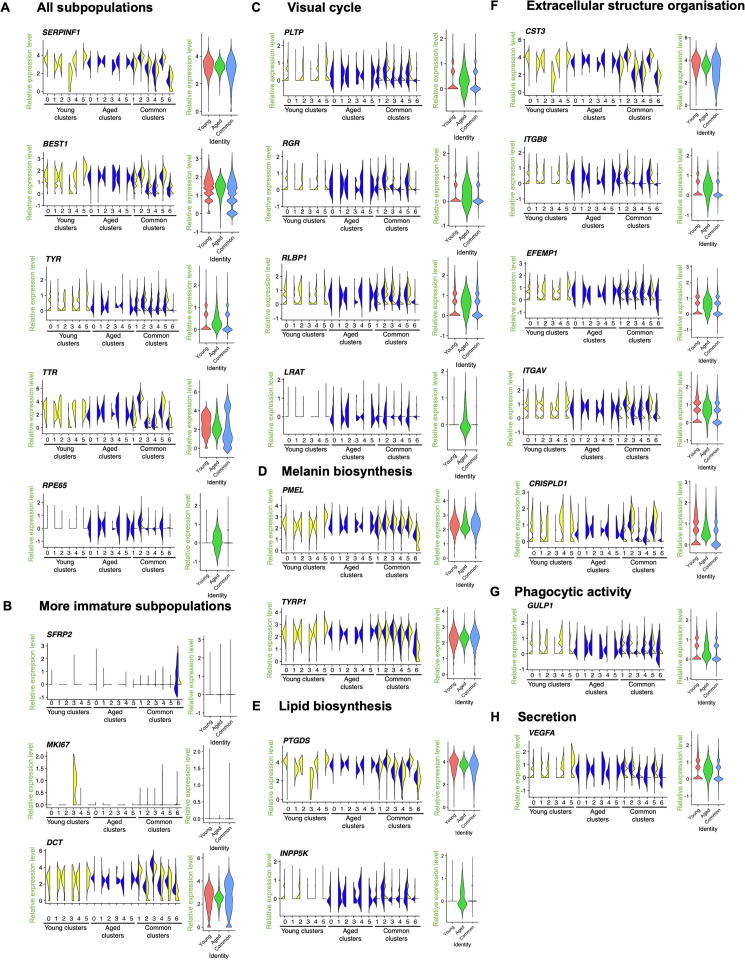
Figure 7.
hPSC-derived RPE subpopulations express native RPE markers with different patterns
Violin plots of selected markers representative of native RPE cells (obtained from [28]) across all subpopulations and in the three main populations “Young”, “Aged”, “Common”, represented in different colours. Subpopulations arising from the 1-month-old culture are indicated in yellow; subpopulations arising from the 12-monht-old culture are indicated in blue. A. Genes found in all subpopulations (SERPINF1, BEST1, TYR, TTR, and RPE65). B. Genes found in more immature subpopulations (SFRP2, MKI67, and DCT). C. Genes with varied expression associated with visual cycle (PLTP, RGR, RLBP1, and LRAT). D. Genes with varied expression associated with melanin biosynthesis (PMEL and TYRP1). E. Genes with varied expression associated with lipid biosynthesis (PTGDS and INPP5K). F. Genes with varied expression associated with extracellular structure organisation (CST3, ITGB8, EFEMP1, ITGAV, and CRISPLD1). G. Genes with varied expression associated with phagocytic activity (GULP1). H. Genes with varied expression associated with secretion (VEGFA). The plots describe the distribution and relative expression of each gene in the subpopulations, measured as normalised UMI counts.