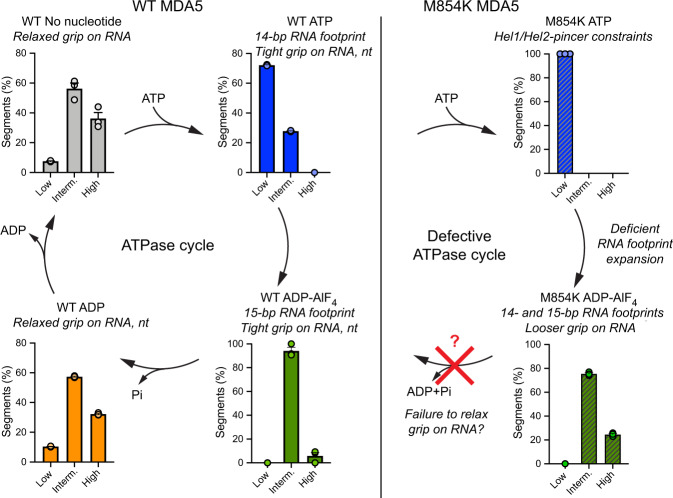
Fig. 6. Helical twist of MDA5 filaments in the ATPase cycle and proposed effect of M854K on the twist.
Histograms showing the distributions of 3D cryoEM reconstructions as a function of helical twist for WT and M854K MDA5-dsRNA filaments with ATP, ADP-AlF4, ADP, or no nucleotide bound. The distributions shown are from 3D classifications performed with ten classes per dataset. Error bars represent SEM between 3D classification calculations, centered on the mean (n = 3 independent calculations). The histograms for nucleotide-free, ATP-bound, and ADP-AlF4-bound WT MDA5-RNA filaments are from data reported previously14. Low, twist = 71˚–81˚; Interm., twist = 81˚–91˚; High, twist = 91˚–96˚. “nt”, ANP nucleotide. See also Supplementary Fig. 8. Source data are provided as a Source Data file.