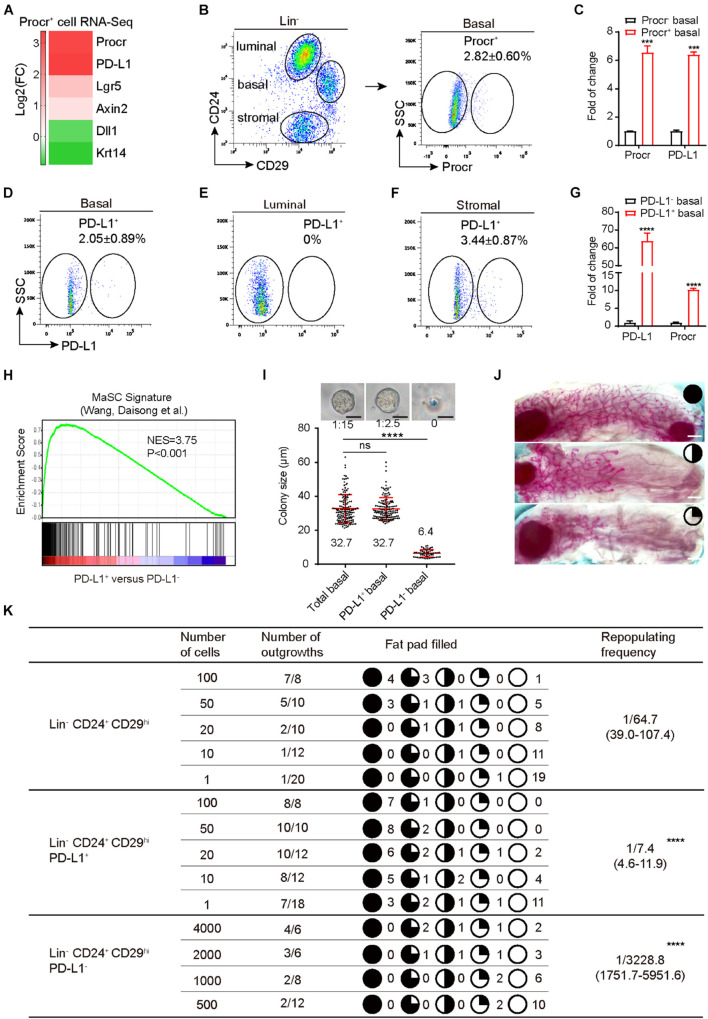
FIGURE 1.
PD-L1 labels a subset of mammary basal cells with high regeneration capacity. (A) PD-L1 expression was enriched in Procr+ MaSCs (Procr+ mammary basal cells). Heat map analysis for Procr+ MaSCs compared with Procr– basal cells. (B) Isolation of Procr+ MaSCs by FACS. (C) Real-time quantification PCR (qPCR) analysis of PD-L1 mRNA levels in Procr+ basal and Procr– basal cells, with GAPDH as an internal control. Data are presented as mean ± SD. Unpaired t-test: *** P < 0.001; n = 3 mice. (D–F) FACS analysis of PD-L1 expression in mammary glands of adult mice. PD-L1+ cells were distributed among basal cells (2.05 ± 0.89%) (D), and stromal cells (3.44 ± 0.87%) (F), no PD-L1+ cells were detected among the luminal cells (E); n = 3 mice. (G) Real-time quantification PCR (qPCR) analysis of Procr mRNA levels in PD-L1+ basal and PD-L1– basal cells, with GAPDH as an internal control. Data are presented as mean ± SD. Unpaired t-test: **** P < 0.0001; n = 3 mice. (H) GSEA demonstrating enriched gene signatures of Procr+ MaSC in the PD-L1+ basal cells as compared to the PD-L1– basal cells. (I) Colony formation efficiency and colony size of total (Lin– CD24+ CD29hi), PD-L1+ (Lin– CD24+ CD29hi PD-L1+) and PD-L1– basal cells (Lin– CD24+ CD29hi PD-L1–) in 3D Matrigel culture. Data were combined from three independent experiments. Data are presented as the mean ± SD. Unpaired t-test: **** P < 0.0001. Scale bars, 50 μm. (J) Representative images of different percentage of fat pad filled with regeneration mammary gland. Scale bars, 1 mm. (K) Regeneration efficiency of total (Lin– CD24+ CD29hi), PD-L1+ (Lin– CD24+ CD29hi PD-L1+) and PD-L1– basal cells (Lin– CD24+ CD29hi PD-L1–). The mammary outgrowth numbers and sizes (shown as the percentage of fat pad filled) are combined from three independent experiments. The mammary reconstitution unit (MRU) frequency is shown for each group. Student’s t-test: **** P < 0.0001. PD-L1, programmed cell death ligand 1; MaSCs, mammary stem cells; Procr, protein C receptor; FACS, fluorescence-activated cell sorting; GSEA, gene set enrichment analysis.