Abstract
Background
MicroRNA (miR)-34a, as a master tumor suppressor in colorectal cancer (CRC), could regulate multiple genes participating in tumor proliferation, invasion, immune evasion, and inflammation-induced progression. Exosomes, as novel nano-carriers, were found to be capable of shuttling crucial mediators to various cells. Since the conventional CRC therapeutics currently are a matter of debate, implication of microRNAs in malignancy remedies have been addressed illustrating promising outlooks.
Objectives
In this study, we aimed to investigate the delivery of miR-34a to CRC cell line CT-26 by encapsulating into tumor-derived exosomes (TEXs), in order to evaluate the anti-proliferative and progressive effects of the novel nano-carrier complex under in vitro condition.
Methods
Exosomes were purified from the starved CT-26 cells and then enriched by miR-34a using the calcium chloride (Cacl2) modified solution. Following the detection of miR-34a expression in the enriched TEXs, the viability of CT-26 cells treated by multiplicity concentrations of either TEXs or TEX-miR-34a was examined. Moreover, the apoptosis rate of the cells was evaluated, and the migration of CT-26 cells subjected to both TEX-miR-34a and TEX was also measured. Thereafter, the expressions of miR-34a target genes, as IL-6R, STAT3, PD-L1, and VEGF-A, which play roles in tumor progression, were determined in the treated CT-26 cells.
Results
The viability of CT-26 cells was harnessed following the treatment with TEX-miR-34a and the apoptosis levels of the cells were also observed to be enhanced dose-dependently. TEX-miR-34a was able to diminish the migration rate of the TEX-miR-34a treated cells and the expressions of IL-6R, STAT3, PD-L1, and VEGF-A were significantly restricted. Moreover, TEXs alone increased the apoptosis rate of tumor cells and repressed the proliferation and migration of these cells which were boosted by enrichment of TEXs with miR-34a.
Conclusion
Exosomes isolated from the starved CT-26 cells were found to have a potential to deliver miR-34a into tumor cells properly with high functionality maintenance for miR-34a in case of regulating genes related to tumor progression and TEXs which showed no positive effect favoring cancer cells, presumably act as a favorable adjuvant in the CRC therapy.
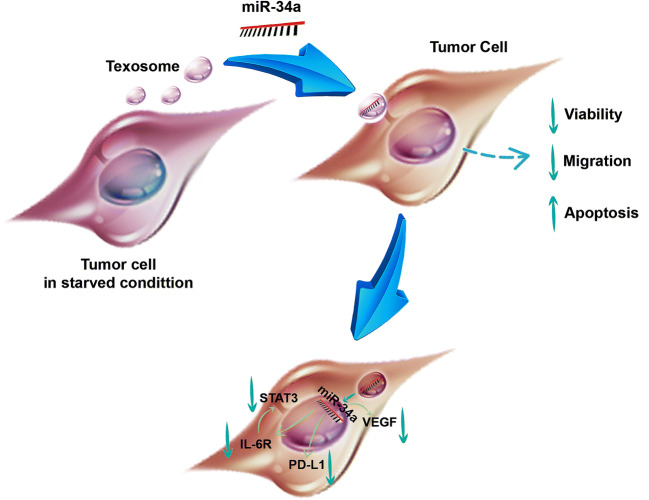
Graphical abstract
Keywords: Texosome, MicroRNA, Colorectal cancer, Apoptosis, Inflammation, Nano delivery
Introduction
Colorectal cancer (CRC) is one of the most controversial cancers, for which routine treatments, including surgery and chemo-radiotherapy, were not satisfying. In this regard, even in many cases, remedy failures have resulted in further invasion and metastasis [1–3]. Thus, in pursue of finding an effective therapeutic option, microRNA (miRNA)-based therapies have recently gained attentions. Correspondingly, miRNAs are small noncoding RNAs, which can regulate the expression of multiple target genes post-transcriptionally, so alterations in their expression levels may substantially affect the fate of tumorgenesis and pathogenesis of diseases [4].
MicroRNA (miR)-34a acts as a tumor suppressor and it can be down-regulated in most malignancies, particularly those tumors related to digestive tract like colorectal cancer [5]. miR-34a can inhibit the progression of cancers through exerting a wide range of impacts on proliferation, invasion, and immune evasion of tumor cells [6]. Indeed, miR-34a was found to be capable of inducing apoptosis in malignant cells [7] as well as modulating cellular migration through targeting vascular endothelial growth factor (VEGF) in CRCs [8]. Moreover, miR-34a was suggested to reduce inflammation by affecting pro-inflammatory cytokine IL-6 secreted by various types of carcinomas and immune cells, particularly at the advanced stages of tumorigenesis. IL-6 is an important factor playing a role in tumor promotion of inflammation through the IL-6R/ STAT3/ miR-34a feedback loop [9, 10]. In this regard, the elevated levels of IL-6 were observed to be related to some factors, including tumor size enlargement, metastasis, developed stages of disease, and declined survival rate of CRC patients [11]. Beside the role of STAT3 in IL-6 signaling pathway and inflammation process, it was indicated to have a critical impact on tumor metastasis, proliferation, cell cycle, and apoptosis, so it was considered as a promising molecular therapeutic target in recent studies [12]. Furthermore, tumor cells express program death ligand 1 (PD-L1), in order to induce exhausted immune cells and to escape from anti-tumor activity of immune system [13], which is related to the reduced miR-34a level in some cancers [14–16]. Additionally, PD-L1 plays a crucial role in protecting tumor cells from cytolysis as well as sustaining self-renewal capacity beyond its function in immune modulation [17–19], which shows substantial roles of PD-L1 in onco-genesis.
Despite performing intensive studies on miRNA interventions in cancer therapy, introducing an effective strategy on delivering miRNAs has been always challenging. Although artificial nano-carriers broadly served in miRNA delivery [20–22], they have been shown to have some drawbacks such as non-uniform particle size distribution, rapid clearance by the reticulo-endothelial system (RES), and toxicity consequences [23–26]. Recently, the possibility of exosomes in the field of drug delivery has made a strong impression in these fields of investigations. Notably, exosomes as small vesicles with size ranged from 30 to 100 nm, are secreted from various cell types. Furthermore, as natural nano-carriers, they can transfer various biological mediators such as proteins, nucleic acids, and RNA species to nearby cells [27]. From a comparative point of view, exosomes possess some favorable features such as biocompatibility, low immunogenicity, low toxicity, biological barrier permeability, and stability in the circulation that have made exosomes promising nano-carriers [28]. In addition, based on the cellular origin of exosomes and surrounding microenvironment, the constituents of exosomal cargoes, which can specifically affect target cells, were found to be varied. For instance, tumor-derived exosomes (TEX) known as texosomes can transfer multiple signaling molecules to nearby cells and this can consequently affect cellular procedures and pathogenesis of cancers [29]. Some collective investigations performed in this field have revealed that in vitro environmental stressors such as heat, hypoxia, and starvation, can increase the secretion and modify the composition of exosomes in various cell types [30–32]. Accordingly, exogenous cargoes can be loaded using various strategies such as electroporation, incubation, vector based transfection, and freeze-thaw treatment [33]. Thus, this study aimed to examine the efficacy of certain suppressing functions of miR-34a in a texosome delivery system isolated from the starved CT-26 murine colon cancer cell line under in vitro condition.
Methods
Cell culture
The mouse colon cancer cell line CT-26 was prepared from Cell Bank of the Pasture Institute of Iran which was verified to be free of mycoplasma contamination. Cells were cultured in RPMI-1640 (Gibco, Paisley, UK) with 100 U/ml of penicillin, 100 mg/ml streptomycin (Gibco, Paisley, UK) and 5 g/l L-glutamine (Gibco, Paisley, UK) which were supplemented by 10% fetal bovine serum (FBS, Gibco, Paisley, UK) and incubated at 37 °C in a 5% CO2 atmosphere. When CT-26 cells grew up to 70–80%, the medium was completely removed and cells were rinsed twice with phosphate-buffered saline (PBS). Then, the cells were grown in serum-free medium [34] and after 24 h post incubation the conditioned medium (CM) was collected.
Isolation and characterization of CT-26 cells derived exosome
CT-26 tumor cell line derived exosome (texosomes;TEX) were isolated from CM using exosome isolation kit (Exocib, Tehran, Iran) according to the manufacturerʼs protocol. Briefly, freshly conditioned medium was pre-cleaned by Eppendorf 5810 R Centrifuge (Eppendorf, Hamburg, Germany) at 300×g for 5 min and 2000×g for 10 min to remove cells and debris contamination. For better result, it was filtered by 0.22 μm filter. Then the supernatant was mixed with reagent A of Exocib kit at ratio 5:1 respectively (the reagent A was vortexed and heated to 37 °C before usage). The mixture was further mixed thoroughly by vortexing for 5 min and incubated at 4 °C overnight. Next, the mixture was centrifuged 40 min at 800×g, supernatant was discarded and the pellet containing exosomes resuspended in 50-200 μl of reagent B or sterile PBS. Finally, the product was aliquot and kept in −80 °C for further use.
Dynamic light scattering (DLS)
The size analysis of texosomes was determined with ZetaSizer Nano series Nano-ZS (Malvern Instruments Ltd., Malvern, UK) at 532 nm with an angle of 90°. Total protein content of texosomes was determined using Bicinchoninic acid assay (BCA, Thermo scientific, Rockford, USA).
Electron microscopy
The morphology of texosomes was visualized by both field emission scanning electron microscope (FESEM) and transmission electron microscope (TEM). For FESEM imaging, the texosome suspension were dried on a glass substrate overnight at room temperature and after gold sputtering directly observed by FESEM instrument (SEM, EM3200, KYKY). For TEM analysis, about 20 μL of texosomes solution was fixed on a copper grid for 5 min. The un-evaporated solution was removed with filter paper. The texosome sample was negatively stained with 1% phosphotungstic acid solution (pH 7.5) for 5 min and measured by TEM (TEM, Philips, Germany).
Western blot
Exosomes were lysed with RIPA lysis buffer (50 mM Tris-HCl, pH 7.5, 1.0 mM EDTA, 150 mM NaCl, 1% Sodium deoxycholate, 0.1% SDS and 1% Triton X-100) with protease and phosphatase inhibitors (Sigma, USA) prior to use. The concentration of proteins was measured using a BCA protein assay kit (Thermo scientific, Rockford, USA) and the protein samples were loaded on SDS-PAGE gel for electrophoresis. Then the proteins were transferred to PVDF membranes (Millipore, USA). Following incubation with primary antibodies (CD9, CD81, CD63 and TSG101) (Abcam, USA) for an overnight, secondary antibodies, goat anti-rabbit horseradish peroxidase immunoglobulin G (diluted 1:10000, System Biosciences, Palo Alto, CA) and goat anti-mouse IgG (diluted 1:2000, Bio-Rad, Watford, UK) were added for next two-hours incubation. Blots were detected using the Chemiluminescence (ECL) system (Santa Cruz, CA, USA). Protein expressions were normalized against β actin expression.
Texosome enrichment by miR-34a
Enrichment of TEXs by miR-34a mimics (AccuTarget™ Custom mouse miRNA mimic, Bioneer, USA) was performed using Cacl2, a modified method of transfection established recently by Duo Zhang et al., [35]. In details, 100 pmol miRNA mimic was mixed with 75 μg texosomes in 150 μl PBS and then Cacl2 (0.1 M concentration) was added. The final volume of mixture was adjusted to 300 μl and incubated on ice for 30 min. Following being heat shocked at 42 °C for 60 s again it was placed on ice for further 5 min. The texosomes were then incubated with 2 μg/ml of RNase A (EN0531, Thermo Fisher) for 20 min at 37 °C to remove all free miRNA mimics in the mixture. Next 2 μg/ml RNase inhibitor (Thermo Fisher) was added for 15 min at 37 °C to inactivate RNase A.
Cellular up-take of miR-34a-enriched texosome
To examine the efficacy of delivery miR-34a-enriched texosome (TEX-miR-34a) by cancer cells, firstly, 5 × 104 CT-26 cells were seeded in 24-well plates and cultured in RPMI-1640 supplemented by 10% FBS overnight. Cells were further treated with 50 μg/ml concentrations of TEX-miR-34a or TEX at 37 °C for 24 h. Following the supernatant was discarded; the cells were harvested to examine miRNA expression by real-time PCR.
Quantitative real-time PCR
Total RNA was isolated using total RNA isolation kit (Yekta Tajhiz Azama, Tehran, Iran) according to the manufacturer’s instructions. In order to analyze mature miRNA, cDNA was generated and qPCR was performed using cDNA synthesis kit (Exiqon, Copenhagen, Denmark), SYBR Green Master Mix, (Ampliqon, Herlev, Denmark) and commercially available primers. For mRNA analyses, cDNA was synthesized from 2 μg total RNA per sample using cDNA synthesis kit (Yekta Tajhiz, Tehran, Iran). qPCR was performed by Rotor-Gene Q system (Qiagen, Hilden, Germany) and the Fast SYBR Green Master Mix (Ampliquin). mRNA and miRNA expression were normalized using detection of housekeeping genes, 60S ribosomal protein L13 and small nu- clear RNA (snRNA) U6 respectively. Results are represented as fold change through the 2-ΔΔCt method with the control set to 1. The sequences of oligonucleotides used as qPCR primers are listed in Table 1 which were designed using the OLIGO v. 7.56 software (Molecular Biology Insights, Inc., USA) or referred to previous studies.
Table 1.
Quantitative Real-time PCR primers used in this study
| Name | Sequence | Ref. |
|---|---|---|
| Mmu-miR-34a-5pa |
Fw: 5´-GCGGCGGTGGCAGTGTCTTAGC-3′ Rv: 5´-ATCCAGTGCAGGGTCCGAGG-3´ |
[36] |
| snRNA U6b |
Fw: 5´-GCTTCGGCAGCACATATACTAAAAT-3′ Rv: 5´-CGCTTCACGAATTTGCGTGTCAT-3´ |
[36] |
| Ribosomal protein L13 |
Fw: 5´-AGCAGATCTTGAGGTTACGGA-3′ Rv: 5´-GGCATGAGGCAAACAGTCT-3´ |
Designed |
| STAT3c |
Fw: 5´-TCGCTCACGTTTGACATGGA-3′ Rv: 5´-TCTAACAACCAACCCCGAGC-3´ |
Designed |
| IL-6Rαd |
Fw: 5´-CGACACTGGGGACTATTTATGC-3′ Rv: 5´-CCCGTTGGTGGTGTTGATT-3′ |
Designed |
| PD-L1e |
Fw: 5´-ACTTGCTACGGGCGTTTAC-3′ Rv: 5´-CTGAAGTTGCTGTGCTGAGG-3´ |
Designed |
| VEGF-Af |
Fw: 5´- CTGCTGTAACGATGAAGCCCTG-3′ Rv: 5′- GCTGTAGGAAGCTCATCTCTCC-3´ |
[37] |
aMouse musculus-microRNA-34a-5p
bSmall nu-clear RNA U6
cSignal Transducer And Activator Of Transcription 3
dInterleukin-6 receptor α
eProgrammed death-ligand 1
fVascular endothelial growth factor A
Cell viability assay
The viability of CT-26 cells was measured using MTT 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay. In details, CT-26 cells (104 cells/well) were seeded in 96-well plates and inoculated with increasing multiplicity concentrations of TEX and TEX-miR-34a for 24 and 48 h. Next, 100 ul of MTT (1 mg/ml, Sigma, USA) was added into each well and incubated at 37 °C. After 4 h, the supernatant was discarded and 50 ul of DMSO (Sigma, USA) was added into each well for 30 min incubation at room temperature. Later, optical density (OD) was taken at 24 and 48 h by a spectrophotometer at 570 nm wavelength. All experiments were performed in triplicate.
Annexin V apoptosis assay
To assay tumor cell apoptosis, an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide apoptosis detection kit (Biolegend, San Diego, CA) was used according to the manufacturer’s instructions. Specifically, CT-26 cells were seeded in 24-well plates, cultured overnight and then treated with various concentrations (10, 25 and 50 μg/ml) of TEX-miR or TEX for 24 h in 500 μl of RPMI-1640 with 10% FBS medium in triplicate. Untreated cells were considered as control. The treated and control cells were harvested and mixed with 5 μl Annexin V-FITC (Biolegend) and 10 μl of 20 μg/ml PI (Biolegend) reagents. The cells were then incubated at room temperature in the dark for 20 min. Then, the samples were subjected to flow cytometry system (FACSCanto, BD, USA) after adding 400 μl PBS, to quantify apoptotic cells through detecting cells positive for Annexin V-FITC.
Wound healing assay
CT-26 cells were seeded in a six-well plate at a density of 2 × 105 cells/well to form a monolayer. The wells at 85–90% confluency were scratched by a 100 μL pipette tip in triplicate. Detached cells were removed and replaced with the media containing moderate concentration (25 μg/ml) of TEX and TEX-miR-34a. Following 24 h, wound closure of scratches were measured through images taken under a phase contrast microscope (Eppendorf, Hamburg, Germany) and image J software (version 1.41o, Java 1.6.0_10, Wayen Rasband, USA).
Statistical analysis
All data were expressed in the form of the mean ± standard deviation (SD). Inter-group comparisons were performed by a Kruskal-Wallis non-parametric analysis of variance and t-test was done to compare two by two analysis. All experiments were performed in triplicate and analyzed using Prism v.8.0 software (GraphPad, La Jolla, CA). Significant statistic difference was defined as p < 0.05.
Results
Characterization of CT-26 cell derived exosomes
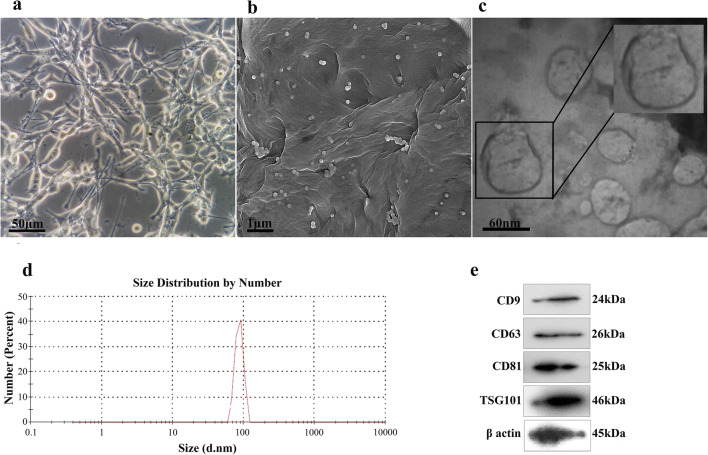
Characteristic analysis illustrated that the average size of TEXs was 87.13 (±10.89) nm with −19.7 ± 12 zeta potential, which was in the reported range of exosome size [36]. As well, the exosome markers, including CD9, CD63, CD81, and TSG101 were verified by immunoblot assay. Furthermore, we assessed the TEXs with electron microscopy and then confirmed the size range of less than 100 nm with spheroid morphology (Fig. 1).
Fig. 1.
Characterization of texosomes. (a) Murine colon cancer cell line CT-26 cultured for texosome isolation, original magnification ×400. (b) Field emission scanning electron microscope (FESEM) image showed spherical shape of isolated texosomes with the diameter less than 100 nm. (c) Transmission electron microscope (TEM) image represents bilayer trans-membrane of CT-26 cell exosomes. (d) The average size of tumor-derived exosomes measured by DLS was 87.13 nm. (e) Surface markers of texosomes which were verified by western blot
High efficiency transfection of miRNA into texosomes
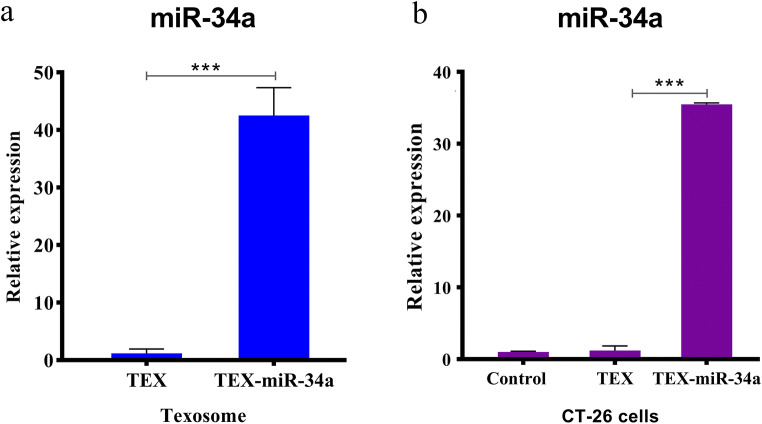
Equal volumes of TEX and TEX-miR-34a were examined to investigate the amount of miR-34a incorporation using real-time PCR. Based on the obtained results, miR-34a expression significantly increased in TEX-miR-34a in comparison with TEX. As well, the fold changes of miR-34a expression enhanced from 1.00 (±0.76) to 42.51 (±4.85) in the enriched texosome following the transfection process (P < 0.001, Fig. 2a). Secondly, the expression of miR-34a was evaluated in the CT-26 cell line co-cultured with 50 μg/ml of TEX or TEX-miR for 24 h. The results showed that the expression level of miR-34a mimic significantly increased in the cells treated with TEX-miR-34a (35.48 ± 0.20) when compared with TEX (1.00 ± 0.10, P < 0.001). This data confirmed the successful delivery of TEX-miR-34a to CT-26 cells. However, no significant difference was detected between the cells treated with TEX and the untreated control cells in miRNA expression (Fig. 2b).
Fig. 2.
Efficacy of loading miR-34a into texosome. (a) The fold changes of miR-34a expression in TEX following transfection and (b) in CT-26 cells treated by TEX or TEX-miR-34a after 24 h incubation. Amplification of PCR was normalized against U6 miRNA (***P < 0.001). TEX: Tumor-derived exosome, TEX-miR-34a: Tumor-derived exosome loaded by miR-34a
miR-34a-loaded TEXs decreased the viability of CT-26 cells
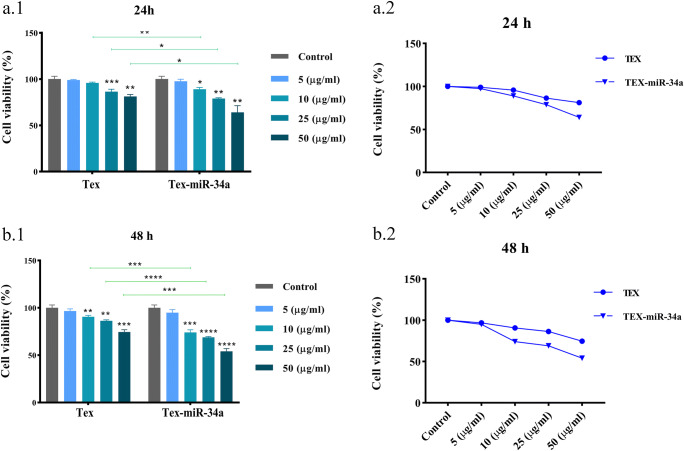
To determine the susceptibility toward TEX-miR-34a during 24 and 48 h, the CT-26 cells were treated by different concentrations of both TEX and TEX-miR-34a. Our results demonstrated that the proliferation rate of cells has significantly inhibited following 24 h by 10, 25, and 50 μg/ml of TEX-miR-34a (88.97% ± 1.89, 78.88% ± 0.88, and 64.0% ± 7.13, respectively) in comparison with the untreated cells (100% ± 3.02, P < 0.05) (Fig. 3a). The inhibitory effect of TEX-miR-34a (10 μg/ml, 73.97% ± 2.76, 25 μg/ml and 68.88% ± 0.88, and 50 μg/ml, 54.03% ± 2.92, respectively) was more significant after 48 h of incubation with tumor cells in compared with the control (100% ± 3.00, P < 0.001) (Fig. 3b). Interestingly, our data depicted that TEX is capable of reducing the CT-26 cell viability, which was significant at both 25 and 50 μg/ml concentrations (86.39% ± 2.63 and 81.12% ± 2.21, respectively) after 24 h (Fig. 3a) as well as in 10, 25, and 50 μg/ml intensity levels (90.53% ± 1.46, 86.25% ± 1.09, and 74.53% ± 2.33, respectively) following 48 h when compared with the controls (100% ± 3.07 and 100% ± 3.04, respectively, P < 0.01) (Fig. 3b).
Fig. 3.
CT-26 cell viability. CT-26 cells were treated with multiplicity density of TEX-miR-34a and TEX for (a.1) 24 h and (b.2) 48 h. Line graph of tumor cells viability following 24 h (a.2) and 48 h (b.2). (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). TEX: Tumor-derived exosome, TEX-miR-34a: Tumor-derived exosome loaded by miR-34a
miR-34a-enriched TEXs induces apoptosis in CT-26 cell line
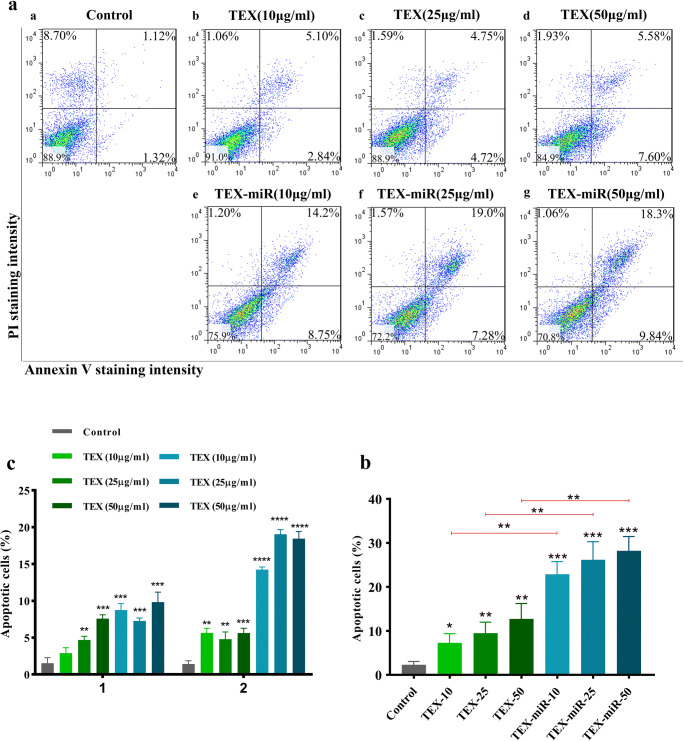
Due to a significant reduction in proliferation rate of cancer cells by TEX-miR-34a after 24 h (Fig. 2a), the following examinations were done during 24 h. As presented in Fig. 3, TEX-miR-34a significantly increased the apoptosis rate of the cancer cells both at early and late apoptosis stages when compared with the untreated cells in a dose-dependent manner (P < 0.001). The rate of apoptosis increased from 2.31% (±0.75) in the untreated cells to 22.90% (±2.87), 26.18% (±4.12), and 28.21% (±3.25) in the treated cells with 10 μg/ml, 25 μg/ml, and 50 μg/ml TEX-miR34a, respectively (Fig. 4a, b). The early and late apoptosis levels also followed the similar increasing manner in tumor cells inoculated by TEX-miR-34a; however, the cells treated with 25 μg/ml showed lower apoptosis levels at early stage and higher ones at late stage (Fig. 4c). Moreover, the elevated levels of apoptotic tumor cells co-cultured by different concentrations of TEX-miR-34a were significant in comparison with the cells treated with the relative concentrations of TEX (P < 0.01, Fig. 4b). Besides, the percentage of apoptotic cells indicated a rise following the treatments with 10 μg/ml (7.31% ± 2.07), 25 μg/ml (9.49% ± 2.50), and 50 μg/ml (12.73% ± 3.54) of TEX compared with the control indicating a similar manner at early stage, but not at late stage of apoptosis (Fig. 4b, c). Given that the concentration of 10 μg/ml of TEX was not significant in both proliferation inhibition and eliciting early apoptosis in tumor cells, the dose of 25 μg/ml was chosen for conducting further tests (Fig. 4c).
Fig. 4.
FITC-Annexin V/PI assay in CT-26 cells. (a) Dot blot and (b) graph showing the apoptosis rate (both early and late apoptosis) of CT-26 colon cancer cell line cultured by TEX or TEX-miR-34a in different concentrations (10, 25 and 50 μg/ml) following 24 h. (c) Early and Late apoptosis in CT-26 cells (*P < 0.05, ** P < 0.01, *** P < 0.001, ****P < 0.0001). TEX: Tumor-derived exosome, TEX-miR: Tumor-derived exosome loaded by miR-34a
TEX-miR-34a declines migration and invasion of CT 26 cells
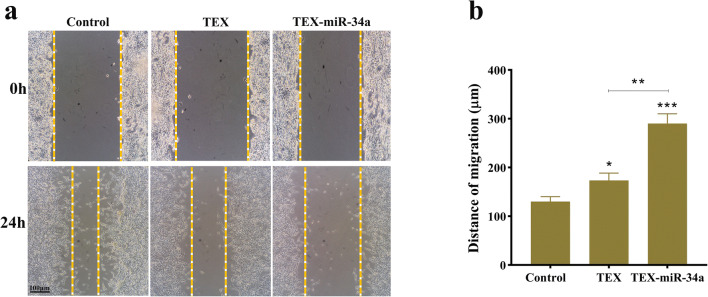
To examine the potential effect of TEX-miR-34a on CT-26 cell migration and invasion, wound healing scratch test was done. As illustrated in Fig. 5, an obvious reduction was observed in the CT 26 migration following the treatment with TEX-miR-34a (130 μm ±10.00) for 24 h compared with the TEX (173.3 μm ±15.28) treatment and untreated cells (290 μm ±20.00, P < 0.01). Notably, tumor cells with TEX treatment showed a significant decrease in their migration compared to the control (P < 0.05).
Fig. 5.
Migration assay. (a) CT-26 cells migration in the presence of TEX and TEX-miR-34a (25 μg/ml) after 24 h. (b) Bar chart of tumor cells migration (*p < 0.05, **p < 0.01, ***P < 0.001). TEX: Tumor-derived exosome, TEX-miR: Tumor-derived exosome loaded by miR-34a
The reduced target gene expressions related to inflammation (IL-6R, and STAT3), tumor invasion (VEGF-A), and immune suppression (PD-L1) by TEX-miR-34a
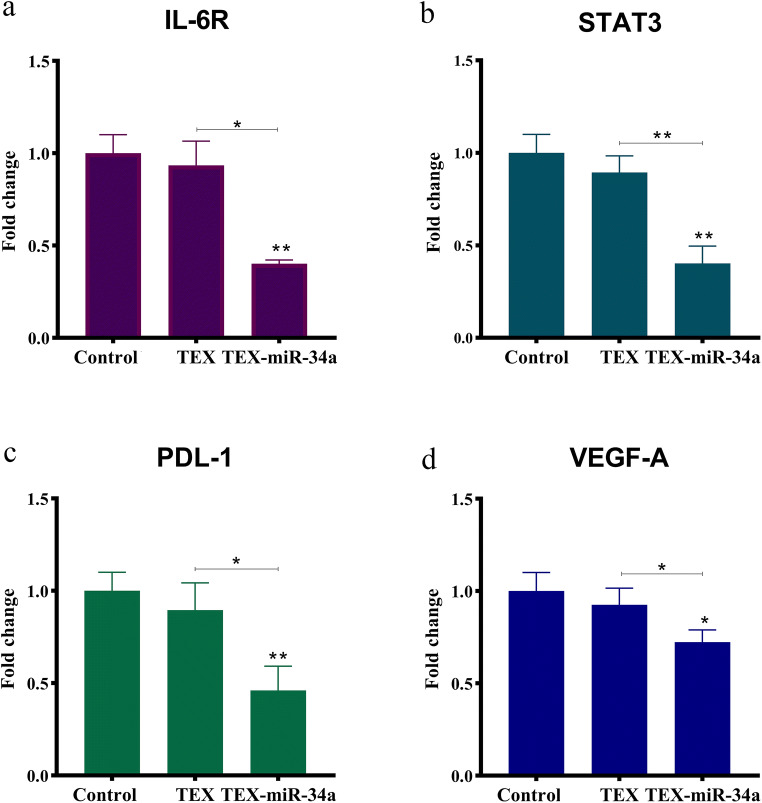
In order to investigate the effectiveness of miR-34a carried by texosome on modulating the expression of some important target genes (IL-6R, STAT3, VEGF-A, and PD-L1), CT-26 cells were treated by a moderate concentration (25 μg/ml) of TEX-miR-34a or TEX for a 24-h duration. As illustrated in Fig. 6, the expression levels of IL-6R transcript significantly decreased in the tumor cells cultured with TEX-miR-34a when compared to the TEX-treated cells and untreated cells (p < 0.05). Additionally, the fold change of IL-6R mRNA decreased from 1.00 ± 0.12 in the untreated cells and 0.93 ± 0.13 in the TEX-treated cells to 0.40 ± 0.01 in the cells treated with TEX-miR-34a. The expression levels of STAT3 significantly decreased in the CT-26 treated by TEX-miR-34a (0.37 ± 0.09) in comparison with the TEX-treated cells (0.89 ± 0.09) and untreated controls (1.00 ± 0.10, p < 0.05). Moreover, the expression levels of PD-L1 showed a significant decrease in those cells co-cultured with TEX-miR-34a from 1.00 ± 0.10 and 0.87 ± 0.14 to 0.47 ± 0.13 in comparison with the untreated cells and the cells treated with TEX (p < 0.05). Additionally, the fold change of VEGF-A mRNA significantly reduced in the cells treated with TEX-miR-34a (0.70 ± 0.11) when compared with both TEX-treated (0.92 ± 0.09) and untreated cells (1.00 ± 0.10, p < 0.05). Interestingly, the expression levels of the target genes for miR-34a showed a decline in those tumor cells treated with TEX; however, these decrease rates were not significant.
Fig. 6.
Relative mRNA expression levels of IL-6R, STAT3, PD-L1 and VEGF-A. The expression levels of IL-6R, STAT3, PD-L1 and VEGF-A were evaluated in CT-26 cell line treated by 25 μg/ml of TEX or TEX-miR-34a transfection following 24 h. Amplification of PCR was normalized against 60S ribosomal protein L13 RNA (*P < 0.05, ** P < 0.01). IL-6R: Interleukin 6 receptor, STAT3: Signal transducer and activator of transcription 3, PD-L1: Programmed death-ligand 1, VEGF-A: Vascular endothelial growth factor-A
Discussion
miRNAs play an extensive role in cancers, and miRNA-based therapies were in the center of attention for many years [37]. Exosomes, as promising vehicles, are capable of shuttling and protecting encapsulated miRNAs from enzymatic degradation and cell-specificity delivering with minimum side-effects and maximum miRNA concentrations per each recipient cell [38–40]. Nevertheless, the strategies proposed for the transfection, in order to make exosomes appropriate for the task of miRNA delivery, are still controversial. In this study, for the first time, we introduced miR-34a into texosome using calcium chloride-mediated transfection, which was recently discovered in a study by Duo Zhang et al. [35]. It is noteworthy that calcium chloride could facilitate miRNAs loading to exosomes conveniently and then limit the requirements of specific laboratory apparatus [35]. Recently, Wei Cheng et al. in their study have applied the novel Cacl2-mediated transfection, in order to load miR-125a/b mimic into exosome derived from jejunum tissue of rats [41]. Our results revealed that the expression of the miR-34a encapsulated in TEX 42.51 folds significantly increased over texosome alone. Likewise, the obtained results in tumor cells following the incubation by exosome-shuttling miR-34a for 24 h represented 35.48 folds elevation in miR-34a expression of the CT-26 cells in comparison to the cells treated with texosome that showed a successful delivery of TEX-miR-34a in the tumor cells. These outcomes are in agreement with those of previous reports on in vitro delivery of miR-15a mimic loaded by exosomes to THP-1 macrophages through Cacl2 transfection [35]. On the contrary, we modified Cacl2-related protocol and detected much higher fold changes in miR-34a expression. Although Duo Zhang et al.’s study presumably is the only research supporting our findings, from a comparative point of view, the research conducted by Asadi rad et al. indicated similar fold changes in the miR-155 expression loaded into TEX using electroporation. Accordingly, this was roughly 25 times higher in the dendritic cells (DCs) treated with TEX-miR-155 in comparison with the cells cultivated by TEX alone [42].
In the next step, we assessed the functionality of TEX-miR34a under in vitro. Our results indicated that TEX-miR-34a can significantly limit the viability of tumor cells and then induce apoptosis in CT-26 cell line in a dose-dependent manner. Xichun Xiao et al. in their study demonstrated that the over-expression of miR-34a could restrict proliferation and promot apoptosis of multiple myeloma cell lines [43]. Similar results were also reported by Chang et al., as the profound induction of cell death has occurred in colon cancer cell line HCT116 [44]. Furthermore, we investigated the effects of the transfected TEX on the expressions of some essential miR-34a-targeted genes in cancer development. Based on the previous findings in this regard, human CRC cells were found to increase the expression of IL-6R in response to IL-6 through the activation of oncogenic transcription factor STAT3 to induce epithelial–mesenchymal transition (EMT) and invasion in primary colorectal tumors as well as in CRC cell line [9, 45]. Matjaz Rokavec et al. in their study proved that miR-34a can adversely regulate the expression of IL-6R in a feed-back loop and can also control oncogenic repercussions of IL-6 [45]. Besides, numerous studies conducted on colitis-associated CRC have asserted that STAT3 signaling is essential for inflammation-induced cancer by the up-regulation of key genes participating in tumor proliferation and survival, particularly β-catenin, which is an oncogenic marker contributing to colorectal carcinogenesis [46, 47]. Our observation is in line with that reported by Matjaz Rokavec et al. in their study [45], and TEX-miR-34a was also able to repress the expression of both IL-6R and STAT3 through the above-mentioned pathway.
Moreover, miR-34a has been identified as an important promoter of immune surveillance in cancers through regulating PD-L1 and sensitizing the PD-L1 overexpressing tumor cells to T lymphocyte killing in acute myeloid lymphoma [48]. Anastasiadou et al. in their study demonstrated that miR-34a reconstitution in EBNA2-transfected diffuse large B cell lymphomas decreased PD-L1 expression as well as enhancing its immunogenicity under in vitro [14]. Our findings regarding the modulation of PD-L1 expression through the TEX-miR-34a treatment supported previous outcomes. Besides, it was indicated that miR-34a is functional enough to inhibit PD-L1 expression significantly. Instead of the immune exhausting function, some tumor-progressive roles, including cancer cell proliferation, chemoresistance, and EMT promotion have been demonstrated for PD-L1 in several studies [18, 49, 50], emphasizing the importance of miR-34a implication in the PD-L1 down-regulation. In addition, miR-34a can inhibit tumor cell migration and invasion by VEGF. Park et al. in their study have outlined that VEGF silencing in gastric cancer cell line NCI-N87 could lead to the decreased proliferation, colony formation, and migration of cancer cells under in vitro [51]. Consistently, our data clearly showed that TEX-miR34a is a potential agent to restrict the expression levels of VEGF-A in the treated CT-26 cells and hampered tumor cell invasion.
Aside from some favorable outcomes regarding the TEX delivering miR-34a into tumor cells, we observed that TEX alone may have the abilities of inducing apoptosis in the CT-26 cells and reducing viability and migration of tumor cells, even though this induction was far lower in comparison with TEX-miR34a. This findings is in contrast with those of previous studies introducing texosomes as a tumor progressive agent playing roles in both proliferation and invasion of cancer cells [52, 53]. These results can be explained by the hypothesis proposed by Johann Mar Gudbergsson et al. [54] concerning the possibility of exosomes as a part of autophagy machinery. It has been assumed that during cellular starvation, the cells could produce and secret autophagosomal vesicles to waste unfavorable biomolecules that are harmful for the cells. As well, due to the similarity between autophagy-related pathways and exosomes, this responsibility may be on exosomes, in order to discard poisonous compounds from the cells [54]. Considering that the secretion of exosomes increases during cellular starvation, exosome release might be known as an autophagy-related response to discharge waste and compensate if degradative autophagy pathways are either overloaded or defected [55, 56]. Therefore, introducing TEX obtained from the starved cells to tumor cells growing under normal condition possibly culminated in apoptosis through texosomal cargos. Moreover, it was shown that delivering miR-34a using these TEXs may exaggerate the phenomenon of apoptosis. Correspondingly, this could be evidenced by a study by Takahashi et al., which has displayed that exosome secretion could relieve cellular stress by transferring damaged DNA and chromosomal fragments in the exosome, in order to prevent the generation of excessive reactive oxygen species [57]. Therefore, the usage of the starved TEXs in cancer therapies is considered as a promising approach not only in terms of miRNA delivery, but also as adjuvant components that may synergistically affect the consequences caused by anti-tumoral therapy. This dual beneficial feature can make the starved texosomes as superior to the conventional texosomes isolated under normal condition leading to tumor progression [58], instead of being a part of cancer therapy. However, the exact signaling pathways involved in tumor suppressing effects of the starved texosomes need further investigations.
Conclusions
It was indicated that miR-34a, as a major tumor suppressor, could efficiently function in texosome delivery system and also harness the propagation and invasion of cancer cells. TEX-miR-34a was also able to impede gene expression associated with immune escape and inflammatory-related progression. Moreover, TEXs, as natural nano-carriers, may be considered as promising vehicles to deliver miRNAs in tumor cells efficiently and present potential adjuvant in cancer treatment strategies along with other therapy approaches isolating the exosomes from the starved tumor cells, which are enriched by harmful substances for tumor cells growing under normal conditions.
Acknowledgments
The authors would like to acknowledge Zuhair Mohammad Hassan for his deliberate advice as well as Mahsa Hajivalili and Ardeshir Abbasi for their kind technical assistance in this project.
Code availability
Not applicable.
Authors’ contributions
MH, DA and ME planned the study, MH, DA and KB designed the methodology, MH and KB conducted the study. MH collected and analyzed the data. MH wrote the report. ME and DA were supervisors, KB was an adviser.
Funding
This work was financially supported by Tarbiat Modares University and the National Animal Modeling Network and In vivo Research, Council for Development of Stem Cell Sciences and Technologies, Vice -Presidency for Science and Technology .grant number 52D/5147.
Data availability
All data achieved during the current study are available by the corresponding author on reasonable request.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
None of the authors has any conflicts of interest to declare.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Davar Amani, Email: amanid@sbmu.ac.ir.
Masoumeh Ebtekar, Email: ebtekarm@modares.ac.ir.
References
- 1.Jun HH, Kwack K, Lee KH, Kim JO, Park HS, Ryu CS, et al. Association between TP53 genetic polymorphisms and the methylation and expression of miR-34a, 34b/c in colorectal cancer tissues. Oncol Lett. 2019;17(5):4726–4734. doi: 10.3892/ol.2019.10092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen R, Pudlarz T, Delattre J-F, Colle R, André T. Molecular targets for the treatment of metastatic colorectal Cancer. Cancers. 2020;12(9):2350. doi: 10.3390/cancers12092350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 4.Babaei K, Shams S, Keymoradzadeh A, Vahidi S, Hamami P, Khaksar R, Norollahi SE, Samadani AA. An insight of microRNAs performance in carcinogenesis and tumorigenesis; an overview of cancer therapy. Life Sci. 2020;240:117077. doi: 10.1016/j.lfs.2019.117077. [DOI] [PubMed] [Google Scholar]
- 5.Akbari A, Majd HM, Rahnama R, Heshmati J, Morvaridzadeh M, Agah S, Amini SM, Masoodi M. Cross-talk between oxidative stress signaling and microRNA regulatory systems in carcinogenesis: focused on gastrointestinal cancers. Biomed Pharmacother. 2020;131:110729. doi: 10.1016/j.biopha.2020.110729. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Y-B, Luo H-P, Shi Q, Hao Z-N, Ding Y, Wang Q-S, et al. miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World journal of gastroenterology: WJG. 2014;20(21):6515. doi: 10.3748/wjg.v20.i21.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiao PF, Yao L, Zeng ZL. Catalpol-mediated microRNA-34a suppresses autophagy and malignancy by regulating SIRT1 in colorectal cancer. Oncol Rep. 2020;43(4):1053–1066. doi: 10.3892/or.2020.7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang D, Zhou J, Dong M. Dysregulation of microRNA-34a expression in colorectal cancer inhibits the phosphorylation of FAK via VEGF. Dig Dis Sci. 2014;59(5):958–967. doi: 10.1007/s10620-013-2983-4. [DOI] [PubMed] [Google Scholar]
- 9.Grivennikov SI, Karin M. Inflammatory cytokines in cancer: tumour necrosis factor and interleukin 6 take the stage. Ann Rheum Dis. 2011;70(Suppl 1):i104–i1i8. doi: 10.1136/ard.2010.140145. [DOI] [PubMed] [Google Scholar]
- 10.Schwitalla S, Ziegler PK, Horst D, Becker V, Kerle I, Begus-Nahrmann Y, Lechel A, Rudolph KL, Langer R, Slotta-Huspenina J, Bader FG, Prazeres da Costa O, Neurath MF, Meining A, Kirchner T, Greten FR. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell. 2013;23(1):93–106. doi: 10.1016/j.ccr.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Knüpfer H, Preiss R. Serum interleukin-6 levels in colorectal cancer patients—a summary of published results. Int J Color Dis. 2010;25(2):135–140. doi: 10.1007/s00384-009-0818-8. [DOI] [PubMed] [Google Scholar]
- 12.Hu F, Li G, Huang C, Hou Z, Yang X, Luo X, et al. The autophagy-independent role of BECN1 in colorectal cancer metastasis through regulating STAT3 signaling pathway activation. Cell Death Dis. 2020;11(5):1–13. doi: 10.1038/s41419-020-2467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O'Donnell E, Chapuy B, Takeyama K, Neuberg D, Golub TR, Kutok JL, Shipp MA. Integrative analysis reveals selective 9p24. 1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116(17):3268–3277. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anastasiadou E, Stroopinsky D, Alimperti S, Jiao AL, Pyzer AR, Cippitelli C, Pepe G, Severa M, Rosenblatt J, Etna MP, Rieger S, Kempkes B, Coccia EM, Sui SJH, Chen CS, Uccini S, Avigan D, Faggioni A, Trivedi P, Slack FJ. Epstein− Barr virus-encoded EBNA2 alters immune checkpoint PD-L1 expression by downregulating miR-34a in B-cell lymphomas. Leukemia. 2019;33(1):132–147. doi: 10.1038/s41375-018-0178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L. Regulatory mechanisms and clinical perspectives of miR-34a in cancer. J Cancer Res Ther. 2014;10(4):805–810. doi: 10.4103/0973-1482.146084. [DOI] [PubMed] [Google Scholar]
- 16.Spina V, Bruscaggin A, Cuccaro A, Martini M, Di Trani M, Forestieri G, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018;131(22):2413–2425. doi: 10.1182/blood-2017-11-812073. [DOI] [PubMed] [Google Scholar]
- 17.Azuma T, Yao S, Zhu G, Flies AS, Flies SJ, Chen L. B7-H1 is a ubiquitous antiapoptotic receptor on cancer cells. Blood, The Journal of the American Society of Hematology. 2008;111(7):3635–3643. doi: 10.1182/blood-2007-11-123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghebeh H, Lehe C, Barhoush E, Al-Romaih K, Tulbah A, Al-Alwan M, et al. Doxorubicin downregulates cell surface B7-H1 expression and upregulates its nuclear expression in breast cancer cells: role of B7-H1 as an anti-apoptotic molecule. Breast Cancer Res. 2010;12(4):1–12. doi: 10.1186/bcr2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almozyan S, Colak D, Mansour F, Alaiya A, Al-Harazi O, Qattan A, et al. PD-L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int J Cancer. 2017;141(7):1402–1412. doi: 10.1002/ijc.30834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SWL, Paoletti C, Campisi M, Osaki T, Adriani G, Kamm RD, Mattu C, Chiono V. MicroRNA delivery through nanoparticles. J Control Release. 2019;313:80–95. doi: 10.1016/j.jconrel.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moritake S, Taira S, Ichiyanagi Y, Morone N, Song S-Y, Hatanaka T, Yuasa S, Setou M. Functionalized nano-magnetic particles for an in vivo delivery system. J Nanosci Nanotechnol. 2007;7(3):937–944. doi: 10.1166/jnn.2007.216. [DOI] [PubMed] [Google Scholar]
- 22.Ruan G-X, Chen Y-Z, Yao X-L, Du A, Tang G-P, Shen Y-Q, et al. Macrophage mannose receptor-specific gene delivery vehicle for macrophage engineering. Acta Biomater. 2014;10(5):1847–1855. doi: 10.1016/j.actbio.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Ferrari M. Beyond drug delivery. Nat Nanotechnol. 2008;3(3):131–132. doi: 10.1038/nnano.2008.46. [DOI] [PubMed] [Google Scholar]
- 24.Hu Y-L, Fu Y-H, Tabata Y, Gao J-Q. Mesenchymal stem cells: a promising targeted-delivery vehicle in cancer gene therapy. J Control Release. 2010;147(2):154–162. doi: 10.1016/j.jconrel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 25.Prete ACL, Maria DA, Rodrigues DG, Valduga CJ, Ibañez OC, Maranhão RC. Evaluation in melanoma-bearing mice of an etoposide derivative associated to a cholesterol-rich nanoemulsion. J Pharm Pharmacol. 2006;58(6):801–808. doi: 10.1211/jpp.58.6.0010. [DOI] [PubMed] [Google Scholar]
- 26.Shubayev VI, Pisanic TR, II, Jin S. Magnetic nanoparticles for theragnostics. Adv Drug Deliv Rev. 2009;61(6):467–477. doi: 10.1016/j.addr.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao S, Li J, Zhang G, Wang Q, Wu C, Zhang Q, Wang H, Sun P, Xiang R, Yang S. Exosomal miR-451a functions as a tumor suppressor in hepatocellular carcinoma by targeting LPIN1. Cell Physiol Biochem. 2019;53:19–35. doi: 10.33594/000000118. [DOI] [PubMed] [Google Scholar]
- 28.Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8(19):4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 30.Brohée L, Demine S, Willems J, Arnould T, Colige AC, Deroanne CF. Lipin-1 regulates cancer cell phenotype and is a potential target to potentiate rapamycin treatment. Oncotarget. 2015;6(13):11264–11280. doi: 10.18632/oncotarget.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He J, Zhang F, Rachel Tay LW, Boroda S, Nian W, Levental KR, Levental I, Harris TE, Chang JT, du G. Lipin-1 regulation of phospholipid synthesis maintains endoplasmic reticulum homeostasis and is critical for triple-negative breast cancer cell survival. FASEB J. 2017;31(7):2893–2904. doi: 10.1096/fj.201601353R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naderi M, Pazouki A, Arefian E, Hashemi SM, Jamshidi-Adegani F, Gholamalamdari O et al. Two triacylglycerol pathway genes, CTDNEP1 and LPIN1, are down-regulated by hsa-miR-122-5p in hepatocytes. Archives of Iranian medicine. 2017;20(3):0-. [PubMed]
- 33.Fu S, Wang Y, Xia X, Zheng JC. Exosome engineering: current progress in cargo loading and targeted delivery. NanoImpact. 2020;100261.
- 34.Liang B, Peng P, Chen S, Li L, Zhang M, Cao D, Yang J, Li H, Gui T, Li X, Shen K. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteome. 2013;80:171–182. doi: 10.1016/j.jprot.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 35.Zhang D, Lee H, Zhu Z, Minhas JK, Jin Y. Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. Am J Phys Lung Cell Mol Phys. 2017;312(1):L110–LL21. doi: 10.1152/ajplung.00423.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chopra N, Dutt Arya B, Jain N, Yadav P, Wajid S, Singh SP, Choudhury S. Biophysical characterization and drug delivery potential of exosomes from human Wharton’s jelly-derived mesenchymal stem cells. ACS omega. 2019;4(8):13143–13152. doi: 10.1021/acsomega.9b01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng X, Cao M, Zhang J, Hu K, Yin Z, Zhou Z, Xiao X, Yang Y, Sheng W, Wu Y, Zeng Y. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35(14):4333–4344. doi: 10.1016/j.biomaterials.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Momen-Heravi F, Bala S, Bukong T, Szabo G. Exosome-mediated delivery of functionally active miRNA-155 inhibitor to macrophages. Nanomedicine. 2014;10(7):1517–1527. doi: 10.1016/j.nano.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.S-i O, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 41.Cheng W, Wang K, Zhao Z, Mao Q, Wang G, Li Q, Fu Z, Jiang Z, Wang J, Li J. Exosomes-mediated transfer of miR-125a/b in cell-to-cell communication: a novel mechanism of genetic exchange in the intestinal microenvironment. Theranostics. 2020;10(17):7561–7580. doi: 10.7150/thno.41802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asadirad A, Hashemi SM, Baghaei K, Ghanbarian H, Mortaz E, Zali MR, Amani D. Phenotypical and functional evaluation of dendritic cells after exosomal delivery of miRNA-155. Life Sci. 2019;219:152–162. doi: 10.1016/j.lfs.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Xiao X, Gu Y, Wang G, Chen S. c-Myc, RMRP, and miR-34a-5p form a positive-feedback loop to regulate cell proliferation and apoptosis in multiple myeloma. International journal of biological macromolecules. 2019;122:526–37. [DOI] [PubMed]
- 44.Chang T-C, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rokavec M, Öner MG, Li H, Jackstadt R, Jiang L, Lodygin D, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. The Journal of clinical investigation. 2015;125(3):1362. doi: 10.1172/JCI81340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, Bateman T, Nebelsiek T, Lundgren-May T, Canli Ö, Schwitalla S, Matthews V, Schmid RM, Kirchner T, Arkan MC, Ernst M, Greten FR. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15(2):91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Grivennikov S, Karin E, Terzic J, Mucida D, Yu G-Y, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, Karin M. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15(2):103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Li J, Dong K, Lin F, Long M, Ouyang Y, Wei J, Chen X, Weng Y, He T, Zhang H. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27(3):443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 49.Alsuliman A, Colak D, Al-Harazi O, Fitwi H, Tulbah A, Al-Tweigeri T, et al. Bidirectional crosstalk between PD-L1 expression and epithelial to mesenchymal transition: significance in claudin-low breast cancer cells. Mol Cancer. 2015;14(1):1–13. doi: 10.1186/s12943-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghebeh H, Barhoush E, Tulbah A, Elkum N, Al-Tweigeri T, Dermime S. FOXP3+ T regs and B7-H1+/PD-1+ T lymphocytes co-infiltrate the tumor tissues of high-risk breast cancer patients: implication for immunotherapy. BMC Cancer. 2008;8(1):1–12. doi: 10.1186/1471-2407-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park J-H, Seo J-H, Jeon H-Y, Seo S-M, Lee H-K, Park J-I, Kim JY, Choi YK. Lentivirus-mediated VEGF knockdown suppresses gastric cancer cell proliferation and tumor growth in vitro and in vivo. OncoTargets and therapy. 2020;13:1331–1341. doi: 10.2147/OTT.S234344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim DH, Park S, Kim H, Choi YJ, Kim SY, Sung KJ, et al. Tumor-derived exosomal miR-619-5p promotes tumor angiogenesis and metastasis through the inhibition of RCAN1. 4. Cancer letters. 2020;475:2–13. doi: 10.1016/j.canlet.2020.01.023. [DOI] [PubMed] [Google Scholar]
- 53.Razzo BM, Ludwig N, Hong C-S, Sharma P, Fabian KP, Fecek RJ, Storkus WJ, Whiteside TL. Tumor-derived exosomes promote carcinogenesis of murine oral squamous cell carcinoma. Carcinogenesis. 2020;41(5):625–633. doi: 10.1093/carcin/bgz124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gudbergsson JM, Johnsen KB. Exosomes and autophagy: rekindling the vesicular waste hypothesis. Journal of cell communication and signaling. 2019:1–8. [DOI] [PMC free article] [PubMed]
- 55.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borràs FE, Breakefield X, Budnik V, Buzas E, Camussi G, Clayton A, Cocucci E, Falcon-Perez JM, Gabrielsson S, Gho YS, Gupta D, Harsha HC, Hendrix A, Hill AF, Inal JM, Jenster G, Krämer-Albers EM, Lim SK, Llorente A, Lötvall J, Marcilla A, Mincheva-Nilsson L, Nazarenko I, Nieuwland R, Nolte-'t Hoen ENM, Pandey A, Patel T, Piper MG, Pluchino S, Prasad TSK, Rajendran L, Raposo G, Record M, Reid GE, Sánchez-Madrid F, Schiffelers RM, Siljander P, Stensballe A, Stoorvogel W, Taylor D, Thery C, Valadi H, van Balkom BWM, Vázquez J, Vidal M, Wauben MHM, Yáñez-Mó M, Zoeller M, Mathivanan S. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10(12):e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo M, Zhao X, Song Y, Cheng H, Zhou R. Nuclear autophagy: an evolutionarily conserved mechanism of nuclear degradation in the cytoplasm. Autophagy. 2016;12(11):1973–1983. doi: 10.1080/15548627.2016.1217381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S, Takasugi M, Watanabe S, Kanemaki MT, Obuse C, Hara E. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8:15287. doi: 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin F, Yin HB, Li XY, Zhu GM, He WY, Gou X. Bladder cancer cell-secreted exosomal miR-21 activates the PI3K/AKT pathway in macrophages to promote cancer progression. Int J Oncol. 2020;56(1):151–164. doi: 10.3892/ijo.2019.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data achieved during the current study are available by the corresponding author on reasonable request.