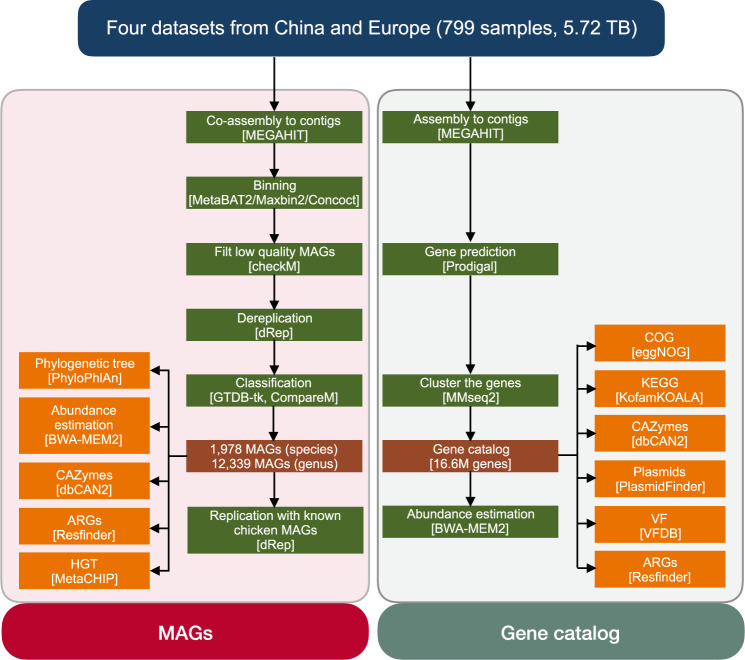
Fig. 1. Flowchart of the steps and bioinformatic tools applied in assembling, constructing, annotating, and analyzing the reference genomes and microbial gene catalog.
Metagenomic sequencing data from the 799 chicken gut microbiome samples from ten countries were integrated. The MAGs (more than 80% completeness and less than 10% contamination) were clustered to strain-level and species-level genome bins at 99% and 95% ANI, respectively. The phylogenetic tree, CAZymes, ARGs, and HGT of the MAGs were analyzed further. The complete genes were clustered to generate the 16.6-million nonredundant gene catalog. Functions of the genes were annotated to profile CAZymes, virulence factors, and plasmid patterns in the chicken gut microbiome.