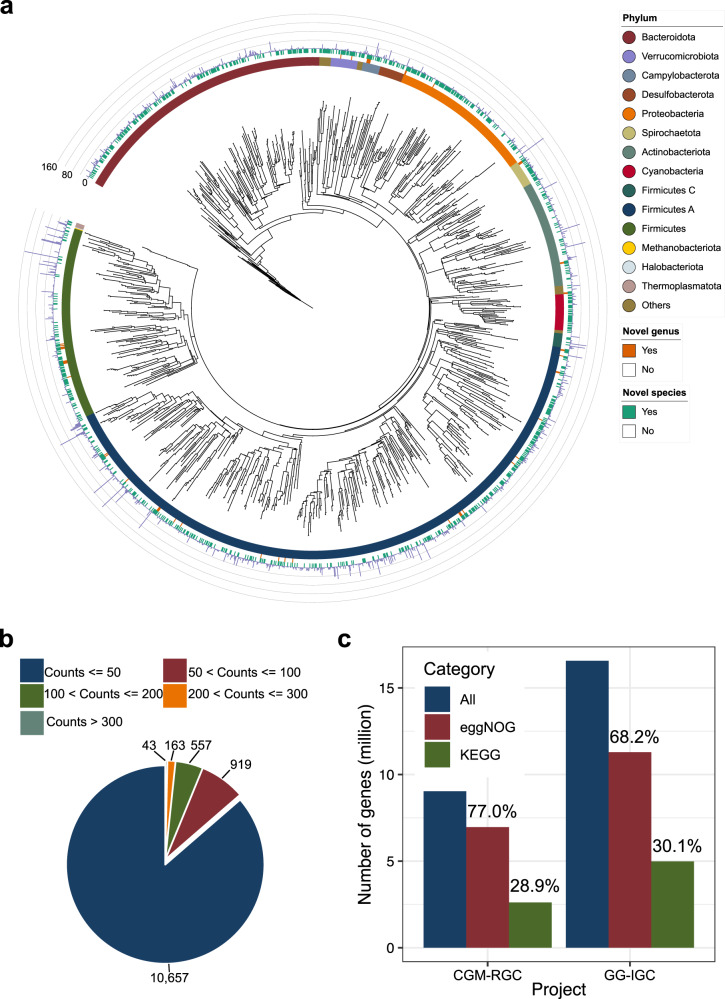
Fig. 2. Metagenome-assembled 12,339 reference genomes and 16,565,684 nonredundant genes from 799 chicken gut microbiomes.
a Phylogenetic tree of 1970 bacterial and eight archaeal species. The taxonomies of the MAGs were assigned by GTDB-Tk. From the inner to outer rings, the first ring represents the phyla, the second ring represents the novel species (n = 893), the third ring represents the novel genera (n = 38), and the height of each bar in the fourth ring represents the number of strain-level genomes in each species. b Distribution of the 12,339 MAGs among gut samples with the criteria of over 1× coverage. For example, 919 MAGs were present in samples with counts between 50 and 100. c Number of genes annotated by eggNOG and KEGG in the CGM-RGC and GG-IGC. The CGM-RGC is a previously published microbial gene catalog of the chicken gut microbiome7. The GG-IGC is the integrated microbial gene catalog produced in the present study.