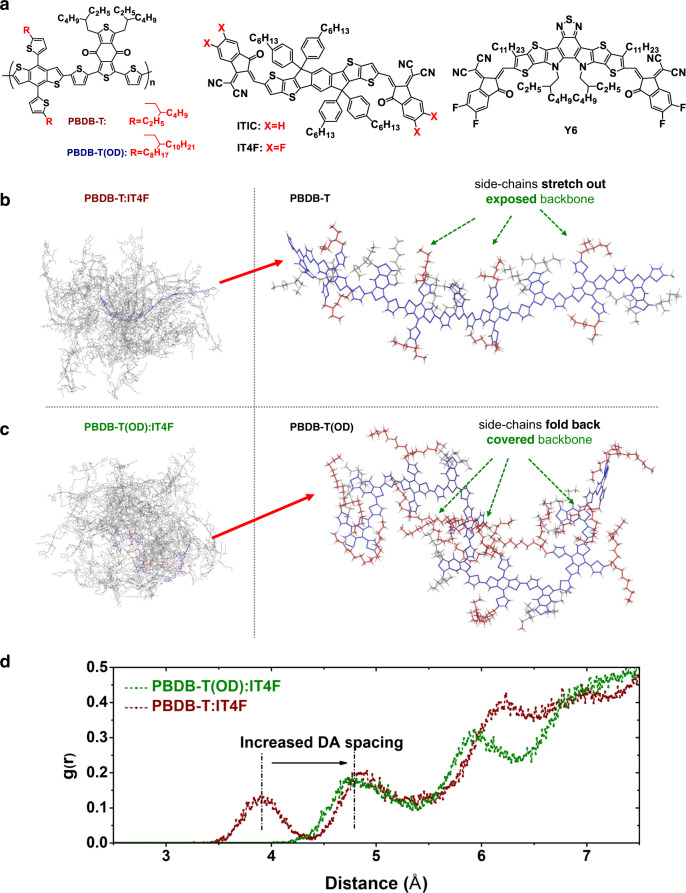
Fig. 3. Molecular dynamic simulations for the BHJ systems.
a Chemical structures of PBDB-T, PBDB-T(OD), and the acceptor materials used. MD simulation results for b PBDB-T and c PBDB-T(OD) blended with IT4F. The side chains of PBDB-T stretch out, leaving the conjugated backbones highly accessible to IT4F, allowing for close parallel π stacking between the backbones and IT4F, while the side chains of PBDB-T(OD) fold back, covering the backbones, reducing the probability for IT4F to stack on the donor backbones. d RDF of the distances between the donor backbones and IT4F for the blends based on PBDB-T and PBDB-T(OD).