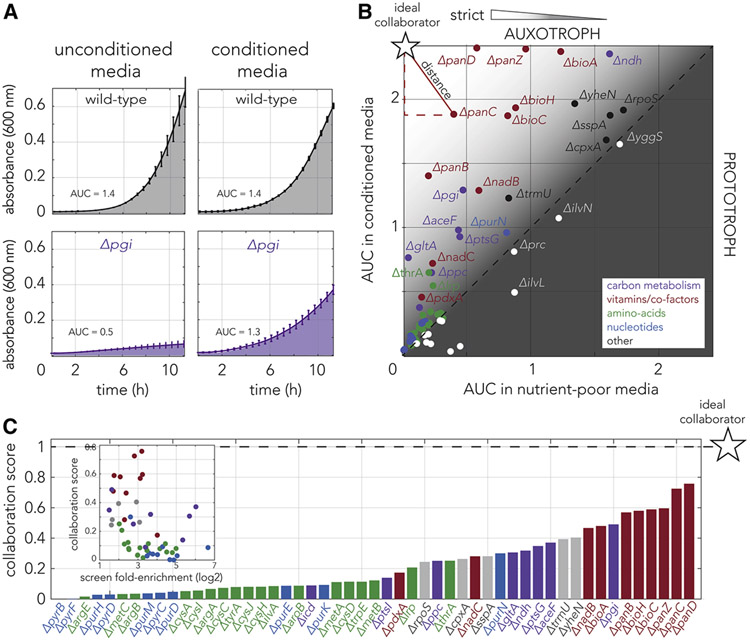
Figure 3. Absolute fitness measurements suggest that strains dysfunctional for specific pathways will be optimal collaborators for cross-feeding interactions.
(A) Growth curves of the wild-type and pgi knockout strains in conditioned and unconditioned media. The area under the curve (AUC), shown as a shaded area, was used as the fitness metric. The fitness of the pgi knockout strain considerably increased on conditioned media. (B) Fitness measurements of 67 knockout strains in conditioned and unconditioned media revealed that strains occupy diverse positions on the fitness plane. The experiment shows that many of the validated hits (strains above the diagonal line) grow reasonably well even in unconditioned media (permissive auxotrophs). The colors mark the biological function of the gene knockout (as in Fig. 2). Knockout strains that are not observed to benefit from conditioned media are marked in white. The star marker indicates the position of a theoretical ideal collaborator, a strict auxotroph that completely arrests on unconditioned media and grows fast on conditioned media. The Euclidean distance from the ideal collaborator, shown as a red segment, was used to calculate the collaboration potential score for each strain. (C) The collaboration optimality score, calculated by each strain’s distance from the ideal collaborator, shows a bias towards some biological functions. The bar graph marks the collaboration optimality score of each knockout strain. Strains scoring high in their collaboration potential are mostly strains dysfunctional for vitamin and co-factor biosynthesis or mutants for carbon metabolism. The inset shows the inverse relationship between the collaboration score and the fold-enrichment measured in the genetic screen.