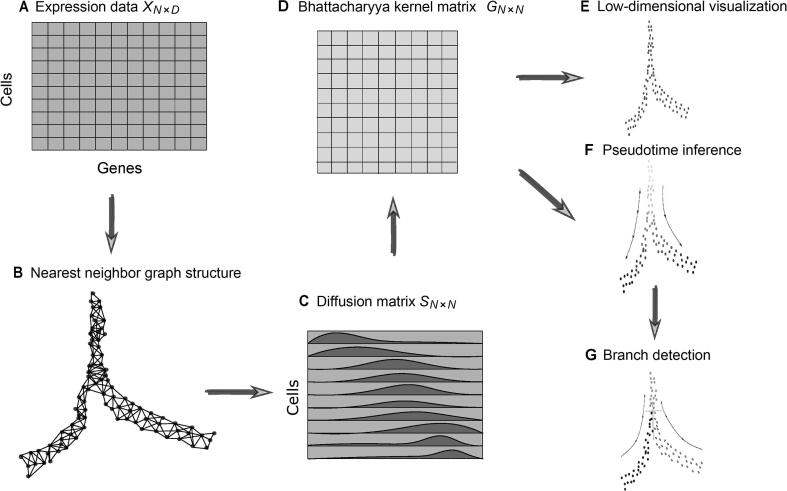
Figure 1.
Overview of DTFLOW algorithm
A. Pre-process a single-cell dataset into a gene expression matrix XN×D with N cells and D genes. B. Compute the nearest neighbors for each cell, get a nearest neighbor graph structure, and then transform the dataset into a Markov transition matrix . C. Use the random walk with restart method to get a diffusion matrix S, in which each cell is represented by a discrete distribution vector. D. Construct a Bhattacharyya kernel matrix and a matrix based on the properties of the kernel method. E. Perform singular value decomposition on to get the low-dimensional embedding . F. Calculate the new distance metric based on the row of the matrix corresponding to the root cell , and unitize it to get the pseudotime distances . G. Identify the multi-branches of cellular differentiation by reverse searching based on the nearest neighbor graph structure.