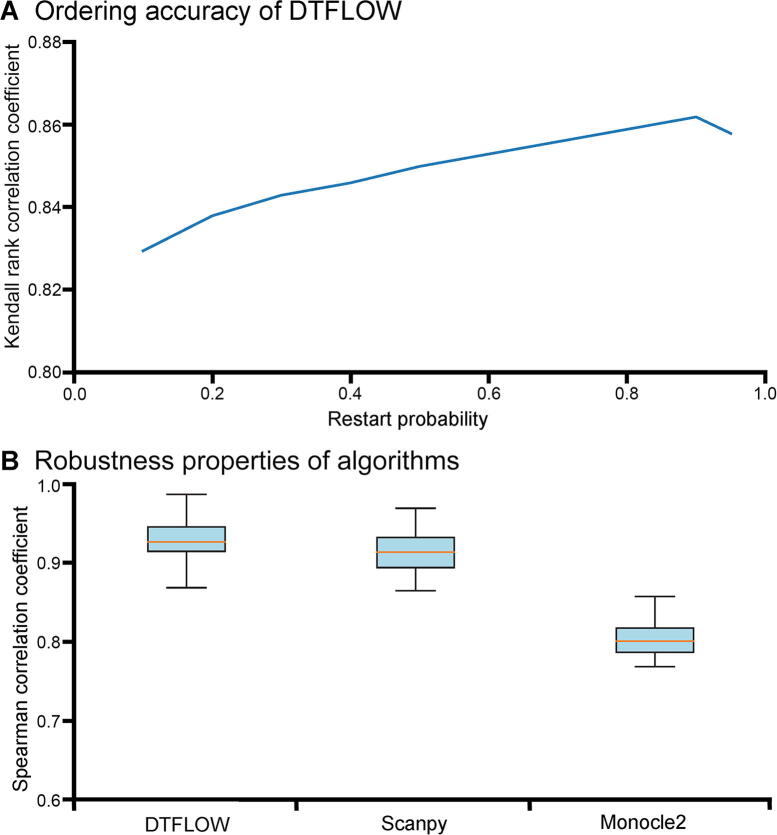
Figure 3.
Accuracy and robustness of three inference methods for the ME dataset
A. The accuracy of DTFLOW determined by different values of restart probability for the ME dataset with 48 genes and 438 single cells [42]. The Kendall rank correlation coefficient is calculated using the stage number of each cell in experimental data and that in the inferred trajectories. B. Mean and standard deviation of the Spearman rank correlation coefficient for three inference methods, including DTFLOW, Scanpy, and Monocle2. The correlation coefficient is calculated using the trajectory of randomly sampled 90% of single cells from the whole dataset and that of the whole dataset. Fifty repeated tests are conducted.