Figure 7. NeuroScope2 - A data viewer for raw and processed extracellular data acquired using multisite silicon probes, tetrodes or single wires.
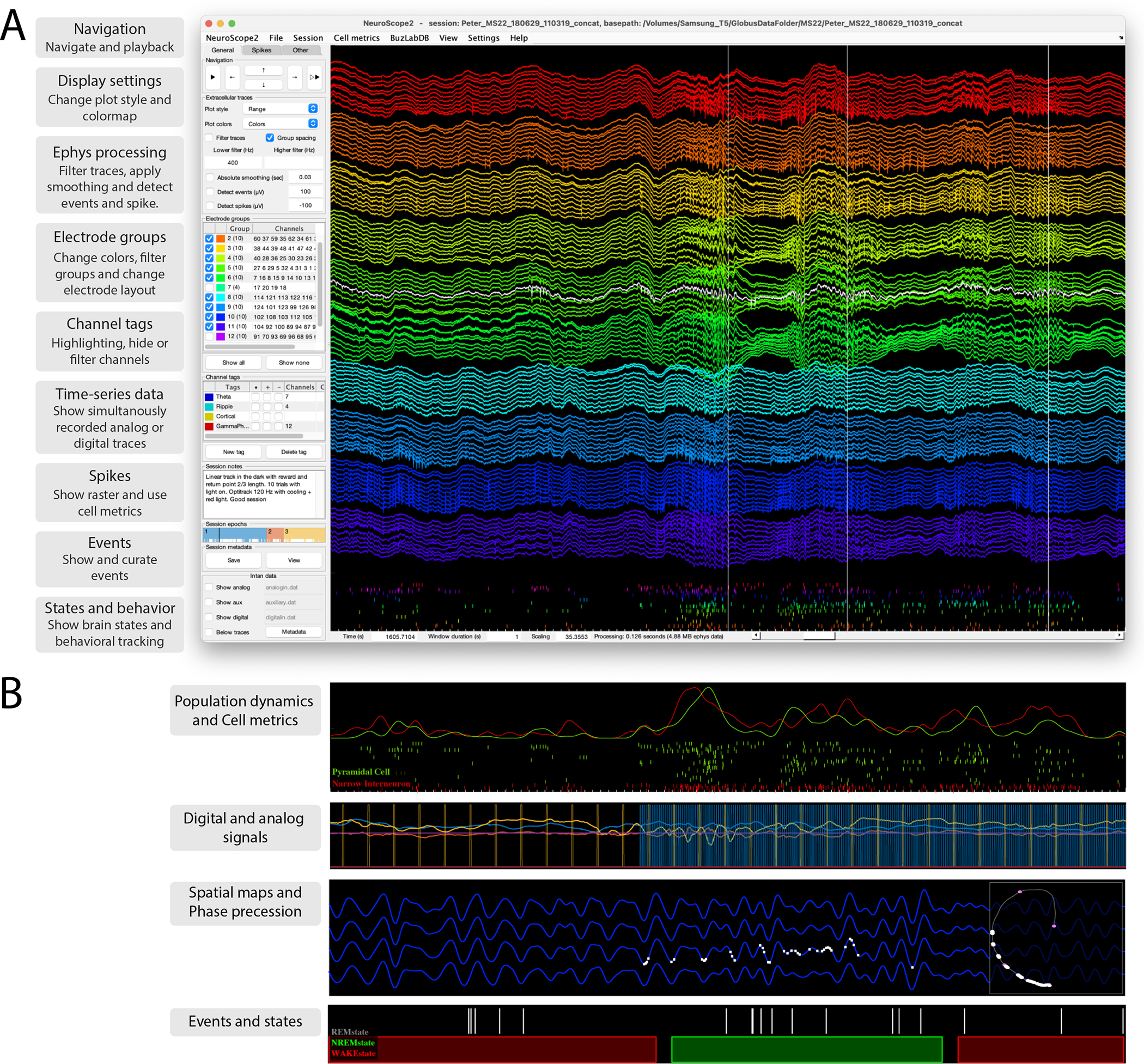
NeuroScope2 is written in Matlab, maintaining many of the original features of NeuroScope (neurosuite.sourceforge.net), but with many enhancements, and it is faster. It is easy to hack or modify, and supports and relies on the data types of CellExplorer. A. Screenshot of the graphical interface, showing a 128-channel recording from the rat hippocampus (window duration = 1 sec). Each colored groups of traces are from the same shank. The three vertical lines are detected temporal events (sharp-wave ripples; detection-channel is highlighted in white). The rasters below the traces are the spikes from curated single units. The left side panel consists of three tabs: General (panels: navigation, ephys traces, electrode groups, channel tags, session notes and epochs, and intan time-series), Spikes (panels: spikes, cell metrics, population dynamics) and Other (panels: events, states, time-series, and behavioral data). B. Various visualizations with NeuroScope2. Top: Population average curves and spiking dynamics of the same population of cells as in A, but color coded and grouped using the putative cell type determined via CellExplorer (duration: 1 sec). Second panel: two digital TTL pulses and 3D accelerometer data (mounted on the animal’s head). Digital data captured using the Intan acquisition system (the TTLs pulses are emitted by a 10Hz camera and a 120Hz behavioral tracking systems; window duration: 3 sec). Third panel: Ephys traces filtered in the theta band, with spikes of a single place cell plotted on the same trace (white bars; window duration: 3 sec), the right square shows the animals spatial trajectory (grey line) and the white points indicate the spatial location of the place field. Lowest panel: Event rater and states data (window duration: 50 sec).
