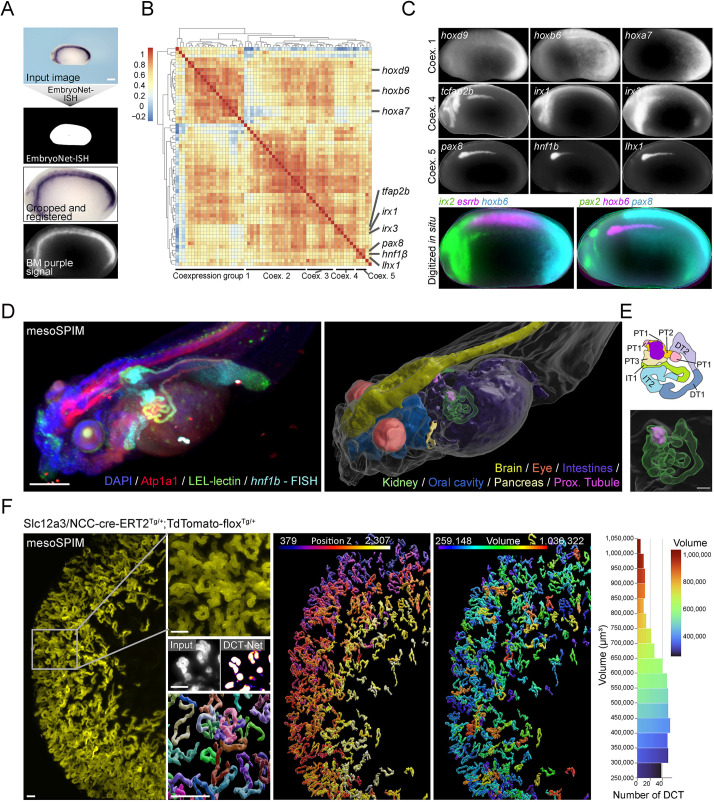
Fig. 6.
Deep learning is applicable to various imaging modalities. (A) EmbryoNet-ISH accurately segmented colorimetric whole-mount in situ hybridization (WISH) stained embryos. (B) Segmentation masks were used to extract, crop and register the in situ signal of stage 26 X. laevis embryos. Unsupervised clustering (n=63) identified distinct co-expression groups. (C) Example images confirmed similar expression patterns within co-expression groups; divergent expression patterns are visualized in multichannel images. (D) mesoSPIM recording of a stage 45 embryo stained for hnf1β using hybridization chain reaction FISH (HCR v3.0), LE-lectin and anti-Atp1a1. DAPI was used as a counterstain. Various morphological structures were segmented in 3D using U-Net models, revealing hnf1β to most strongly expressed in the proximal tubular segments PT2/3 (purple label) and pancreas. (E) Schematic and enlarged view of the pronephros segmentation. (F) mesoSPIM recording of the adult kidney of an induced Slc12a3/NCC-cre-ERT2Tg/+:TdTomato-floxTg/+ reporter mouse to visualize the distal convoluted tubules (DCT). DCT-Net segmented single DCTs and maintained separation of DCTs in close proximity. DCT-Net 3D-segmentations highlighted the spatial distribution of DCT in the renal cortex and permitted feature extraction such as volume measurements. Scale bars: 200 μm (white); 40 μm (gray).