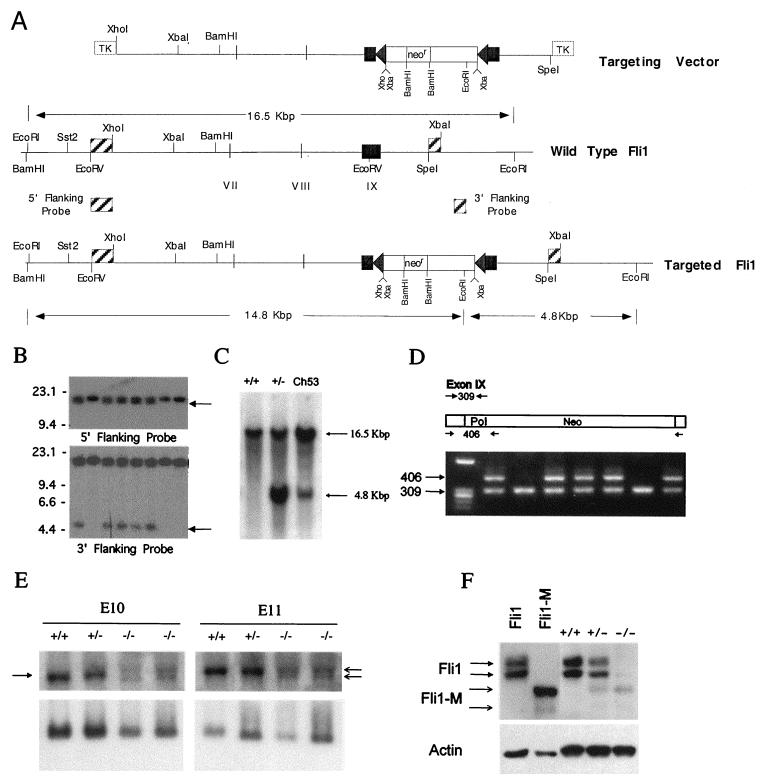
FIG. 1.
Targeted disruption of the mouse Fli1 gene. (A) Schematic representation of the Fli1 targeting vector and genomic organization of the wild-type and Fli1 targeted alleles. The partial restriction map of the mouse Fli1 locus, indicating the positions of exons VII, VIII, and IX, is given. Triangles on each side of the Neo cassette (neor) represent loxP sequences. Positions of diagnostic EcoRI sites and predicted fragment sizes are indicated. (B) Identification of Fli1 targeted cell lines. Southern blot analysis of G418-resistant ES cell clones, probed with the indicated 5′ or 3′ probe, is shown. Targeted ES cell clones are identified by the presence of either a 14.8-kbp (5′ probe) or 4.8-kbp (3′ probe) fragment in addition to the 16.5-kbp fragment from the nontargeted allele. (C) Germ line transmission of the targeted Fli1 allele. Genomic DNA was isolated from wild-type and heterozygous mutant mice and from a chimeric mouse (C53). Southern blot analysis of EcoRI-digested DNA from representative wild-type (+/+) and heterozygous mutant (+/−) and from chimera 53 (Ch53), probed with the 3′ Fli1 probe (described in the legend to Fig. 1A), is shown. (D) PCR analysis of tail DNA from the progeny from a heterozygous intercross. Primers (Fli9F, Fli9R, and Pol IIR) were used to amplify genomic DNA, and the resultant products were resolved on a 1% Trevigel. The wild-type allele is identified by the 309-bp PCR product, and the targeted allele is identified by the 406-bp PCR product. Heterozygous mice are identified by the presence of both the 309- and 406-bp bands. The wild-type mice are identified by the presence of only the 309-bp band. No homozygous mutant mice (identified by the presence of only the 406-bp band) were detected. (E) The Fli1 mRNA transcript level is reduced in mutant embryos. Total RNA was extracted from wild-type and heterozygous and homozygous mutant E10 and E11 embryos. RNA (5 μg/lane) was electrophoresed on 1.2% agarose containing formaldehyde, transferred to a nylon membrane, and sequentially hybridized with 32P-labeled Fli1 cDNA (top panel) and S26 rRNA probes (bottom panel). The arrow on the left highlights the position of the normal Fli1 transcript, and the arrows on the right indicate the position of the two transcripts from the targeted Fli1 allele. (F) Presence of a truncated Fli1 protein in mutant E11 embryos. Total protein (5 μg/lane) prepared from wild-type and heterozygous and homozygous mutant E11 embryos was separated by polyacrylamide gel electrophoresis and transferred to filters. The filters were incubated with a Fli1 polyclonal antibody and visualized using the enhanced chemiluminescence system. Protein prepared from cells after transient transfection with vectors expressing Fli1 or mutant Fli1 (Fli1-M) were used as controls. Stripped membranes were reprobed with β-actin antibody. The 48-kDa protein band in the −/− embryos cannot be wild-type protein, because no wild-type Fli1 transcript is made in these −/− embryos. Furthermore, we did not observe the larger p51 Fli1 protein, normally encoded by the wild-type mRNA utilizing an alternative translation start. The identity of this protein remains unknown but is likely to be a cross-reactive protein.