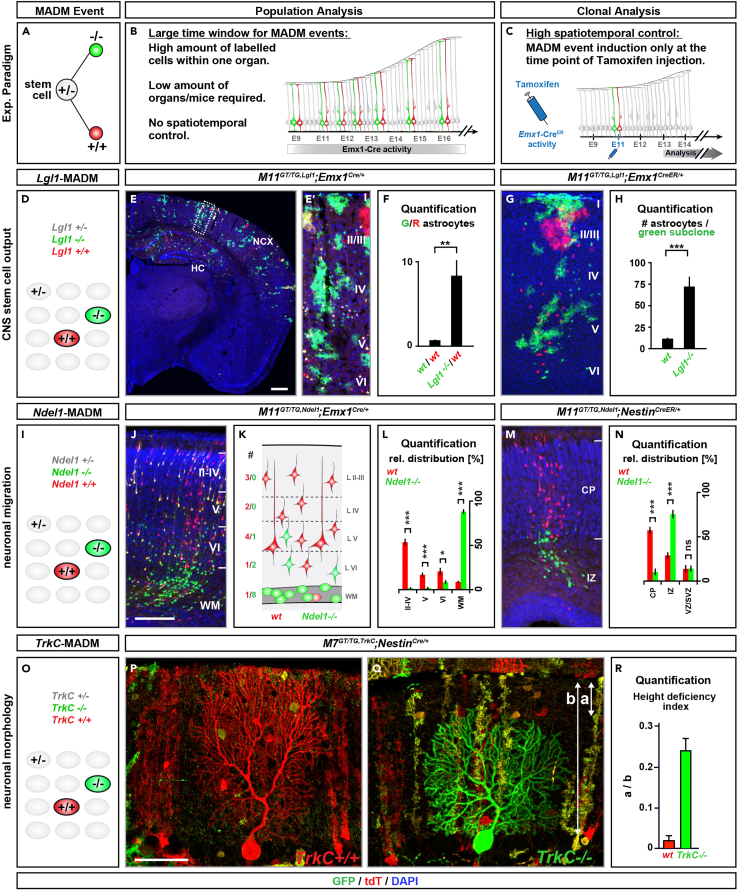
Figure 5.
Examples and analysis of genetic mosaic MADM animals with mosaicism in the brain
(A–C) Schematic of G2-X MADM events resulting in (A) genetic mosaic tissue using (B) constitutively active Cre driver for population analysis and (C) tamoxifen-inducible CreER driver for clonal analysis.
(D–H) Analysis of central nervous system (CNS) stem cell output in Lgl1-MADM mice using Lgl1 mutant allele (Klezovitch et al., 2004). (D) schematic of Lgl1-MADM cortical tissue; (E and F) population analysis of Lgl1 using Emx1-Cre driver (Gorski et al., 2002) with (E) overview of a representative brain section at P21, (E′) insert showing the cortex, and (F) quantification of the G/R ratio of astrocytes from wild-type mice and Lgl1-MADM mice; (G and H) clonal analysis of Lgl1 using Emx1-CreER driver (Kessaris et al., 2006) with (G) reconstruction of a representative clone at P21 and (H) quantification of the number of green astrocytes in green subclones for wild-type and Lgl1 mutant clones. Scale bars: (E) 500 μm, (E′) 60 μm, (G) 70 μm). Values represent mean ± SEM. ns, nonsignificant, ∗∗∗p < 0.001. Parts of the figure and its legend were adapted and reused from (Beattie et al., 2017) with permission.
(I–N) Analysis of cortical projection neuron migration in Ndel1-MADM mice using Ndel1 knock-out allele (Sasaki et al., 2005). (I) schematic of Ndel1-MADM cortical tissue; (J–L) population analysis of Ndel1 using Emx1-Cre driver (Gorski et al., 2002) with (J) overview of a representative image of the cortex section at P1, (K) schematic indicating partitioning of the cortex into layers and layer-specific quantification of red and green neurons, and (L) quantification of the relative distribution of red wild-type and green Ndel1 mutant cells in the respective layers; (M and N) clonal analysis of Ndel1 using Nestin-CreER driver [line 1 in (Imayoshi et al., 2006)] with (M) reconstruction of a representative clone at E18.5 and (N) quantification of the relative distribution of red wild-type and green Ndel1 mutant cells in the respective layers. Scale bars: (J) 150 μm, (M) 100 μm. Values represent mean ± SEM. ns, nonsignificant; ∗p < 0.05 and ∗∗∗p < 0.001. Parts of the figure and its legend were adapted and reused from (Hippenmeyer et al., 2010) with permission.
(O–R) Analysis of Purkinje cell morphology in TrkC-MADM mice using TrkC knock-out allele (Tessarollo et al., 1997) and Nestin-Cre driver (Petersen et al., 2002). (O) schematic of TrkC-MADM neuronal tissue; (P and Q) representative high magnification images of (P) red wild-type and (Q) green TrkC mutant Purkinje cells in the cerebellum of P21 TrkC-MADM mice, (R) quantification of the height deficiency index of red wild-type and green TrkC mutant cells within the Molecular layer of the cerebellum. Scale bars: (P and Q) 50 μm. Presented morphology values are means ± SEM. ∗∗∗ p < 0.001, one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test. Parts of the figure and its legend were adapted and reused from (Joo et al., 2014) with permission.