Abstract
A Gram-stain-negative, obligatory anaerobic spirochaete (RCC2812T) was isolated from a faecal sample obtained from an individual residing in a remote Amazonian community in Peru. The bacterium showed highest 16S rRNA gene sequence similarity to the pig intestinal spirochete Treponema succinifaciens (89.48 %). Average nucleotide identity values between strain RCC2812T and all available Treponema genomes from validated type strains were all <73 %, thus clearly lower than the species delineation threshold. The DNA G+C content of RCC2812T was 41.24 mol%. Phenotypic characterization using the API-ZYM and API 20A systems confirmed the divergent position of this bacterium within the genus Treponema . Strain RCC2812T could be differentiated from the phylogenetically most closely related T. succinifaciens by the presence of alkaline phosphatase and α -glucosidase activities. Unlike T. succinifaciens , strain RCC2812T grew equally well with or without serum. Strain RCC2812T is the first commensal Treponema isolated from the human faecal microbiota of remote populations, and based on the collected data represents a novel Treponema species for which the name Treponema peruense sp. nov. is proposed. The type strain is RCC2812T (=LMG 31794T=CIP 111910T).
Keywords: faeces, gut, human, microbiome, spirochaete, Treponema
Bacteria from the genus Treponema are typically anaerobic, spiral-shaped, highly motile micro-organisms that are fastidious to culture [1]. This genus comprises a number of primary pathogens responsible for syphilis [2] and periodontal disease [3] in humans, as well as digital dermatitis [4] in cattle. Commensal treponemas have received considerably less attention, although they are commonly found in the gastrointestinal tract of some insects [5] and mammals such as pigs and cows [6, 7], as well as most primates [8] including humans [9–12]. Next-generation sequencing data have shed more light on the taxonomic diversity and distribution of autochthonous Treponema members of the human gut microbiota, uncaptured by culturing methods. Treponema species have been consistently recognized as members of the faecal microbiome of humans living a traditional lifestyle, remote from industrialization, across continents, climates [11–13] as well as in extinct human populations (ancient humans from Mexico) [14]. To reveal any key functional role associated to these conserved gut microbes, characterization of cultured representatives is essential.
A previous cross-sectional metagenomic exploration revealed that Treponema was one of the dominant members of the gut microbiome of remote Matsés tribe populations living in small settlements along the rivers of the Peruvian Amazon. Subsequent genome reconstructions from that population suggested that these commensal Treponema strains fell outside the known pathogenic clades and were more similar to Treponema succinifaciens [15]. Here we report on strain RCC2812T, recovered from a human stool sample as part of the Flora Intestinal Nativa project, an integrated study of the effect of industrialization on the gut microbiota of Peruvian populations (unpublished). On the basis of the information obtained from a polyphasic taxonomic approach, we propose that RCC2812T represents a novel species within the genus Treponema .
Isolation and ecology
Strain RCC2812T was isolated from a human stool sample, collected from a seemingly healthy 29-year-old male resident of the Remoyacu village in the Peruvian Amazonian jungle, in the framework of the Flora Intestinal Nativa project (unpublished). The stool sample was collected within 30 min after defecation and stored in a 5 ml tube with 50 % (v/v) of glycerol and frozen immediately at −80 °C. It was transported to our lab on dry ice and further preserved at −80 °C. 16S rRNA gene sequence analysis revealed that the sample was highly enriched in Treponema (30 % of relative abundance). A sample aliquot of 200 mg (wet weight) was enriched in oral Treponema enrichment broth (OTEB; Anaerobe Systems) containing 10 % foetal bovine serum (FBS; Gibco), 25 µg ml−1 rifampin (Sigma-Aldrich) and 5 µg ml−1 enrofloxacin (Sigma-Aldrich) for 24 h at 37 °C in an anaerobic chamber (N2/H2/CO2, 80 : 10 : 10, 37 °C), as previously described [16–18]. The enriched culture was diluted in OTEB and plated on fastidious anaerobe agar (FAA; Lab M), supplemented with 5 % defibrinated sheep blood (Sanbio), 25 µg ml−1 rifampin and 5 µg ml−1 enrofloxacin. Single colonies were picked after 48–72 h, sub-cultured in the broth described above and selected for further characterization based on spiral shape cell morphology, determined using a phase contrast microscope. Isolates were preserved at −80 °C in OTEB broth supplemented with 10 % (v/v) FBS, 5 % (v/v) dimethyl sulfoxide and 5 % (v/v) glycerol. Selected isolates were also subjected to alkaline lysis followed by thermal lysis (15 min at 95 °C), 16S rRNA was then amplified by PCR using AccuPrime Taq DNA polymerase (Invitrogen) with primers fD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and rP2 (5′-ACGGCTACCTTGTTACGACTT-3′) [19]. Primer rP2 was used for Sanger sequencing (Eurofins Genomics). The sequences obtained were taxonomically assigned using NCBI blast. One isolate tentatively allocated to the genus Treponema , RCC2812T, was further characterized taxonomically.
16S rRNA gene phylogeny
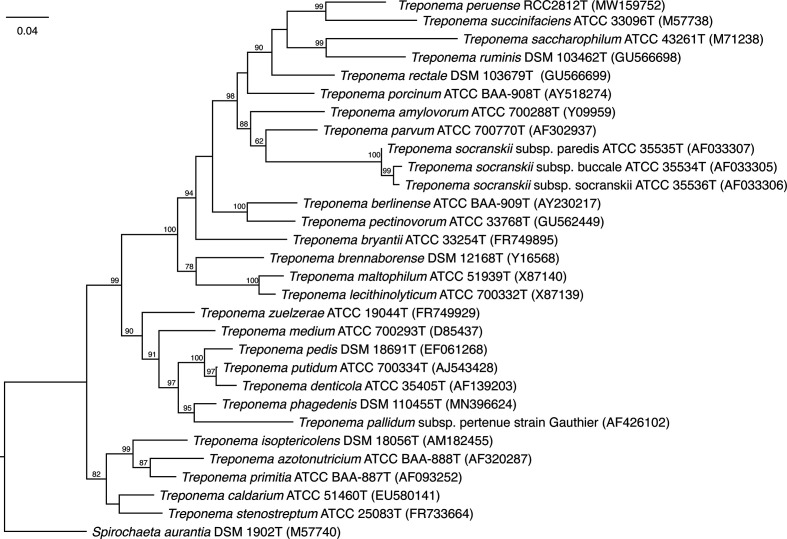
High molecular weight DNA was isolated from RCC2812T using the Puregene cell and tissue kit (Qiagen) according to the manufacturer’s instructions. A standard SMRTbell library with 10 Kb insertions was generated, after which PacBio long reads were sequenced on the PacBio Sequel System (Pacific Biosciences) with SMRTbell Template Prep Kit 1.0-SPv3 (Pacific Biosciences). Library construction and sequencing was performed at Novogene. Sequencing reads were assembled into one circular chromosome representing the RCC2812T genome with coverage >400×. In order to more accurately determine the evolutionary relationships of strain RCC2812T with other Treponema species, we retrieved the 16S rRNA gene identified in its genome sequence. Four copies of the 16S rRNA gene were found in three distinct operons across the genome [1007688–1 169 243(+); 2239131–2240673(−) and 2467010–2468552(−)], they all share above 99 % sequence identity. We performed a search of the 16S rRNA genes against the EzBioCloud 16S rRNA gene database [20], which identified T. succinifaciens DSM 2489T as the closest species with a similarity of 89.48 % (for all four copies of the 16S rRNA gene of strain RCC2812T). muscle alignment of the 16S rRNA genes of all Treponema type strains along with one of the 16S rRNA copies of RCC2812T (1167707–1169238_DIR+=accession MW159752) allowed a FastTree maximum-likelihood phylogenetic tree reconstruction (Fig. 1). This analysis suggested that isolate RCC2812T probably represents a novel Treponema species most closely related to T. succinifaciens and falls within a clade of commensal Treponema originating from the gastro-intestinal tract of different mammalian hosts. Similar results were obtained using the neighbour-joining method (see Fig. S1, available in the online version of this article).
Fig. 1.
Phylogenetic analysis of 16S rRNA genes from all species of the genus Treponema presently recognized along with isolate RCC2812T. This phylogenetic tree was generated with FastTree using the maximum-likelihood method, based on the generalized time-reversible model (there were a total of 1242 nucleotide positions in the final dataset). Bootstrap values (only those >70 % are shown) based on 1000 replicates are shown at the branch nodes for the maximum-likelihood method. GenBank accession numbers are indicated in parentheses. Bar: 0,04 nucleotide substitutions per site.
We further confirmed that strain RCC2812T was a novel species by computing the orthologous average nucleotide identity (OrthoANI [21]) between the genome of RCC2812T and the genome of the other Treponema species whenever available (see Table S1 for a list of all analyses performed on Treponema type strains). The ANI was consistently <73 % against RCC2812T across all pairwise comparisons, the highest being 72.2 % with T. succinifaciens (see Table S2), well below the 95 % threshold for species delineation. Similarly, genome-to-genome distance calculation [22] from the DSMZ culture collection (http://ggdc.dsmz.de/) corroborated that RCC2812T was a new species, with all estimated DNA–DNA hybridization (DDH) values being below 23 % (see details in Table S3).
Genome features
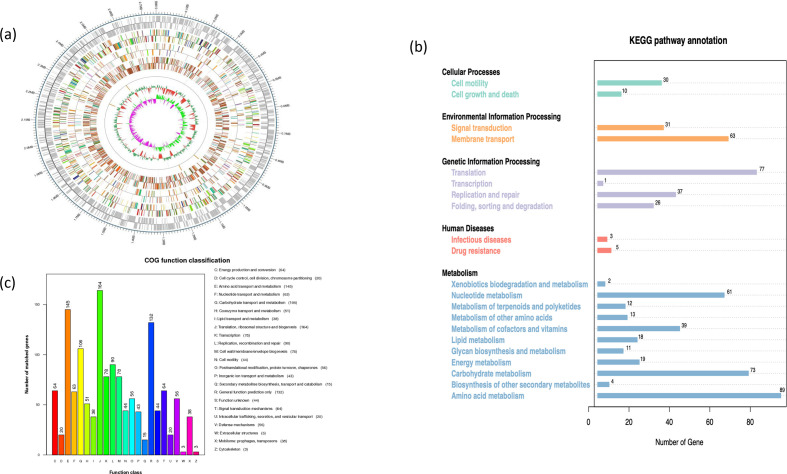
The genome of RCC2812T comprises one chromosome of 2 738 066 bp, with a G+C content of 41.24 mol%, 2580 predicted genes and no detected plasmids. As a reminder, the closest type strain T. succinifaciens DSM 2489T possesses one chromosome of 2 897 425 bp with a G+C content of 39.13 mol%, 2786 predicted genes and a plasmid of 165 572 bp [23]. Out of the 2580 predicted genes of RCC2812T only 1190 have a match in the genome of Treponema succinifaciens DSM 2489T according to the NCBI-blast non-redundant (nr) database), 2033 were functionally annotated using the NCBI-blast non-redundant database (version 201611), 1919 using KEGG (version 201801) and 1294 using COG (see details in Fig. 2).
Fig. 2.
(a) Genome representation of strain RCC2812T mapping from the outside to the inside of the circle: all predicted genes, cog, kegg, go, non-coding RNA, G+C content and G+C skew, (b) gene predictions with COG and (c) KEGG.
To determine if genomes similar to strain RCC2812T have been identified in other metagenomic datasets, we computed the average nucleotide identity of RCC2812T against all the Treponema species genomes available in the Unified Human Gastrointestinal Genome collection [24]. We identified 53 genomes with an average nucleotide identity above 95 %, indicating this species is present in other metagenomic datasets (see Table S4). Interestingly, this species of Treponema was only identified in datasets from remote and rural unindustrialized populations: in Latin America (seven from a rural village in El Salvador [25], 29 from Peru [15, 25], Africa (five from rural communities in Madagascar [26], seven from Tanzania [13]) and Oceania (five from rural agrarian communities in the Fiji Islands [27], having the highest prevalence in Peru (around 11 % of samples).
Morphology and physiology
Cells of strain RCC2812T stain Gram-negative and appear, under a phase contrast microscope, as highly motile with a helical coil. Using electron microscopy (see Fig. 3), RCC2812T exhibited all typical cell morphology features of the genus: small, helical spirochaete with four periplasmic flagella in a 2 : 4 : 2 arrangement. Cells were approximately 4–6 µm long and had a diameter of 0.2–0.3 µm with two to five spirals.
Fig. 3.
(a and b): Conventional scanning electron microscopy images of RCC2812T (a, in spiral shape, b, in cystic form). (c): Transmission electron microscopy images of RCC2812T with negative staining (arrow showing one of the two periplasmic fibrils originating subterminally from the end of the protoplasmic cylinder). (d) Conventional transmission electron microscopy image of a thin side section of a RCC2812T cell (arrow showing again two periplasmic fibrils in bundle surrounded by the outer sheath).
RCC2812T displays good growth on FAA plates supplemented with 10 % FBS and 5 % defibrinated sheep blood at 37 °C under anaerobic conditions, but grows equally well without serum or blood. On the contrary, T. succinifaciens can grow on FAA plates without blood but grows poorly without serum. After 48 to 72 h on FAA, strain RCC2812T produces punctiform translucent non-pigmented colonies with a diameter of approximately 0.5 mm, smooth surface and a slight cream colour. No local haemolysis was observed after 4 weeks of plate culture. The strain reaches stationary phase in unsupplemented OTEB within 72 h. The strain is mesophilic, with growth temperatures ranging from 20–37 °C. Strain RCC2812T is catalase-negative.
Further phenotypic characterization using the API ZYM system (bioMérieux) confirmed that strain RCC2812T represents a phenotypically distinct species within the genus Treponema (see Table 1). The API ZYM test inoculum was prepared by harvesting cells from three fully grown FAA plates supplemented with 10 % FBS and 5 % sheep blood incubated for 48 h. API ZYM test series were determined four times following the manufacturer’s instructions. Of all tested activities, only alkaline phosphatase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase and α-glucosidase activities were positive. As API ZYM data for the phylogenetically closest species T. succinifaciens DSM 2489T were not previously reported, this strain was also included in the API ZYM test series. Strain DSM 2489T exhibited esterase lipase (C8), acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, β-glucosidase and slight α-fucosidase activities; all other reactions were negative.
Table 1.
Comparison of enzyme activity profiles of oral and gastrointestinal Treponema type strains from human, porcine and bovine hosts using the API ZYM system
Enzymes tested: 1, alkaline phosphatase; 2, C4 esterase; 3, C8 esterase lipase; 4, C14 lipase; 5, leucine arylamidase; 6, valine arylamidase; 7, cystine arylamidase; 8, trypsin; 9, chymotrypsin; 10, acid phosphatase; 11, naphtholphohydrolase; 12, α-galactosidase; 13, β-galactosidase; 14, β-glucuronidase; 15, α-glucosidase; 16, β-glucosidase; 17, N-acetyl-β-glucosaminidase; 18, α-mannosidase; 19, α-fucosidase. +, Positive; −, negative.
|
Species/subspecies |
Strain |
Presence of enzyme activity |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 |
||
|
Treponema peruense |
RCC2812T |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
+ |
− |
+ |
− |
− |
− |
− |
|
DSM 2489T |
− |
− |
+ |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
+ |
− |
− |
+ |
− |
− |
−/+ |
|
|
Treponema rectale* |
DSM 103679T |
− |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
|
DSM 103462T |
− |
− |
+ |
− |
+ |
− |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
+ |
− |
− |
− |
|
|
ATCC 700770T |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
+ |
− |
− |
− |
− |
− |
|
|
ATCC BAA-909T |
− |
− |
− |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
|
|
ATCC BAA-908T |
− |
+ |
− |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
+ |
− |
− |
− |
− |
|
|
DSM 18691T |
− |
+ |
+ |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
|
|
ATCC 700293T |
+ |
+ |
+ |
− |
+ |
− |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
− |
− |
− |
− |
|
|
DSM 12168T |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
+ |
− |
+ |
− |
+ |
− |
− |
|
|
ATCC 33768T |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
|
|
Treponema socranskii subsp. Socranskii¶ |
ATCC 35536T |
+ |
+ |
− |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
|
Treponema socranskii subsp. Buccale¶ |
ATCC 35534T |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
+ |
− |
− |
− |
− |
− |
|
Treponema socranskii subsp. Paredis¶ |
ATCC 35535T |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
|
Treponema maltophilum¶ |
ATCC 51939T |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
+ |
+ |
− |
− |
+ |
− |
− |
− |
+ |
|
ATCC 700288T |
+ |
+ |
− |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
− |
+ |
|
|
ATCC 35405T |
− |
+ |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
|
|
ATCC 700334T |
+ |
+ |
+ |
− |
+ |
− |
− |
+ |
+ |
+ |
+ |
+ |
+ |
− |
+ |
+ |
− |
− |
− |
|
|
ATCC 700332T |
+ |
+ |
+ |
− |
− |
− |
− |
− |
− |
+ |
+ |
− |
+ |
+ |
− |
− |
+ |
− |
+ |
|
In addition, strain RCC2812T was also compared to T. succinifaciens DSM2489T using API 20A and API rapid ID32 test series (bioMérieux) following the manufacturer’s instructions. Bacterial inocula were prepared by harvesting cells from three FAA fully grown plates per strain supplemented with 10 % FBS and 5 % sheep blood after 48 h of incubation. In API 20A, strain RCC2812T was positive for acidification of d-glucose, lactose, sucrose, maltose and d-mannose. On the contrary, T. succinifaciens DSM2489T was also positive for acidification of maltose but not of d-glucose, lactose, sucrose and instead it was positive for d-xylose and cellobiose.
Using API rapid ID32, RCC2812T was positive for β-galactosidase, β-galactosidase-6-phosphate and α-arabinosidase. T. succinifaciens DSM 2489T shared only β-galactosidase activity with RCC2812T. Contrary to RCC2812T, T. succinifaciens DSM 2489T had in addition α-glucosidase and β-glucosidase activities.
To determine the main fermentation products of strain RCC2812T, a few colonies were harvested from FAA plates supplemented with 10 % FBS and incubated in unsupplemented OTEB broth for 48 h. The culture was then centrifuged for 10 min at 2000 r.p.m. The resulting supernatant was used for HPLC analysis using Aminex HPLC columns (Bio-Rad Laboratories). T. succinifaciens DSM2489T was used for reference, and OTEB medium as negative control. The major fermentation products of both Treponema strains were formate and acetate, followed by succinate and lactate that were produced in smaller amounts.
Description of Treponema peruense sp. nov.
Treponema peruense (pe.ru.en’se. N.L. neut. adj. peruense pertaining to Peru, where the sample from which the novel species was isolated came from).
Cells are Gram-stain-negative and highly motile under phase contrast microscopy. Under electron microscopy, cells appear as small, helical spirochaetes with four periplasmic flagella in a 2 : 4 : 2 arrangement, 4–6 µm long and with a diameter of 0.2–0.3 µm, and with two to five spirals. Strain RCC2812T is a strict anaerobe that reaches stationary phase in unsupplemented OTEB medium after 72 h. When grown on FAA plates supplemented with 10 % FBS at 37 °C under anaerobic conditions for 48 h, it produces punctiform translucent non-pigmented colonies with a diameter of approximately 0.5 mm, smooth surface and a slight cream colour. In the API-ZYM system, strain RCC2812T exhibited the following enzymatic activities: alkaline phosphatase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase and α-glucosidase. In API 20A assays, strain RCC2812T tested positive for acidification of d-glucose, lactose, sucrose, maltose, d-mannose, raffinose and l-arabinose. Major fermentation products formed in OTEB medium were formate, acetate, succinate and lactate. The genome of this strain comprises one chromosome of 2 738 066 bp, with a G+C content of 41 .24 mol%, 2580 predicted genes and no detected plasmid. On the basis of polyphasic taxonomic data presented here, we suggest recognition of RCC2812T as a novel species within the genus Treponema for which the name Treponema peruense is proposed. The type strain is RCC2812T (=LMG 31794T=CIP 111910T) and was isolated from a faecal sample obtained from an individual residing in a remote Amazonian community in Peru.
Supplementary Data
Funding information
Fonds Wetenschappelijk Onderzoek – Vlaanderen: Research funding FWO G0B7320N, C.B. is funded by FWO through a PhD fellowship (no. 11ZF416N), R.T.T. is funded by FWO through a postdoctoral fellowship (no. 1234321 N), R.B. is funded by FWO through a postdoctoral fellowship (no. 1221620 N). Katholieke Universiteit Leuven, Rega Institute for Medical Research, Vlaams Instituut voor Biotechnologie.
Acknowledgements
We would like to thank the Peruvian participants, specially the Matsés community from Remoyacu, the personnel at the medical post in Colonia Angamos, the field team of the project Flora Intestinal Nativa, the Peruvian NIH and DIRESA (Regional Health Direction) Loreto. We would also like to thank the Electron Microscopy (EM) platform of the VIB Bioimaging Core Leuven for their help with the EM pictures of strain RCC2812T as well as Kristel Bernaerts lab for their contribution to the metabolic profiling.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The protocol for this study was reviewed and approved by the Ethics Committee of the Peruvian National Institute of Health (protocol OEE-010-15), who provided periodic monitoring during the execution of the project.
Footnotes
Abbreviations: ANI, average nucleotide identity; DDH, DNA–DNA hybridization; FAA, fastidious anaerobe agar; FBS, foetal bovine serum; OrthoANI, orthologous average nucleotide identity; OTEB, oral Treponema enrichment broth.
Four supplementary tables and one supplementary figure are available with the online version of this article.
References
- 1.Staton GJ, Newbrook K, Clegg SR, Birtles RJ, Evans NJ, et al. Treponema rectale sp. nov., a spirochete isolated from the bovine rectum. Int J Syst Evol Microbiol. 2017;67:2470–2475. doi: 10.1099/ijsem.0.002051. [DOI] [PubMed] [Google Scholar]
- 2.Forrestel AK, Kovarik CL, Katz KA. Sexually acquired syphilis: Historical aspects, microbiology, epidemiology, and clinical manifestations. J Am Acad Dermatol. 2020;82:1–14. doi: 10.1016/j.jaad.2019.02.073. [DOI] [PubMed] [Google Scholar]
- 3.Asai Y, Jinno T, Igarashi H, Ohyama Y, Ogawa T. Detection and quantification of oral treponemes in subgingival plaque by real-time PCR. J Clin Microbiol. 2002;40:3334–3340. doi: 10.1128/JCM.40.9.3334-3340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demirkan I, Carter SD, Winstanley C, Bruce KD, McNAIR NM, et al. Isolation and characterisation of a novel Spirochaete from severe virulent ovine foot rot. J Med Microbiol. 2001;50:1061–1068. doi: 10.1099/0022-1317-50-12-1061. [DOI] [PubMed] [Google Scholar]
- 5.Schnorr SL, Hofman CA, Netshifhefhe SR, Duncan FD, Honap TP, et al. Taxonomic features and comparisons of the gut microbiome from two edible fungus-farming termites (Macrotermes falciger; M. natalensis) harvested in the Vhembe district of Limpopo, South Africa. BMC Microbiol. 2019;19 doi: 10.1186/s12866-019-1540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cwyk WM, Canale-Parola E. Treponema succinifaciens sp. nov., an anaerobic spirochete from the swine intestine. Arch Microbiol. 1979;122:231–239. doi: 10.1007/BF00411285. [DOI] [PubMed] [Google Scholar]
- 7.Newbrook K, Staton GJ, Clegg SR, Birtles RJ, Carter SD, et al. Treponema ruminis sp. nov., a spirochaete isolated from the bovine rumen. Int J Syst Evol Microbiol. 2017;67:1349–1354. doi: 10.1099/ijsem.0.001812. [DOI] [PubMed] [Google Scholar]
- 8.Manara S, Asnicar F, Beghini F, Bazzani D, Cumbo F, et al. Microbial genomes from non-human primate gut metagenomes expand the primate-associated bacterial tree of life with over 1000 novel species. Genome Biol. 2019;20:299. doi: 10.1186/s13059-019-1923-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Obregon-Tito AJ, Tito RY, Metcalf J, Sankaranarayanan K, Clemente JC, et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat Commun. 2015;6:6505. doi: 10.1038/ncomms7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angelakis E, Bachar D, Yasir M, Musso D, Djossou F, et al. Treponema species enrich the gut microbiota of traditional rural populations but are absent from urban individuals. New microbes new Infect. 2019;27:14–21. doi: 10.1016/j.nmni.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rampelli S, Schnorr SL, Consolandi C, Turroni S, Severgnini M, et al. Metagenome sequencing of the hadza hunter-gatherer gut microbiota. Curr Biol. 2015;25:1682–1693. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 14.Tito RY, Knights D, Metcalf J, Obregon-Tito AJ, Cleeland L, et al. Insights from characterizing extinct human gut microbiomes. PLoS One. 2012;7:e51146. doi: 10.1371/journal.pone.0051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obregon-Tito AJ, Tito RY, Metcalf J, Sankaranarayanan K, Clemente JC, et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat Commun. 2015;6:6505. doi: 10.1038/ncomms7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stamm L, Bergen HL, Shangraw KA. Natural rifampin resistance in Treponema spp. correlates with presence of N531 in RpoB rif cluster i. Antimicrob Agents Chemother. 2001;45:2973–2974. doi: 10.1128/AAC.45.10.2973-2974.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans NJ, Brown JM, Demirkan I, Murray RD, Vink WD, et al. Three unique groups of spirochetes isolated from digital dermatitis lesions in UK cattle. Vet Microbiol. 2008;130:141–150. doi: 10.1016/j.vetmic.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Evans NJ, Brown JM, Murray RD, Getty B, Birtles RJ, et al. Characterization of novel bovine gastrointestinal tract Treponema isolates and comparison with bovine digital dermatitis treponemes. Appl Environ Microbiol. 2011;77:138–147. doi: 10.1128/AEM.00993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parola P, Roux V, Camicas JL, Baradji I, Brouqui P, et al. Detection of ehrlichiae in African ticks by polymerase chain reaction. Trans R Soc Trop Med Hyg. 2000;94:707–708. doi: 10.1016/s0035-9203(00)90243-8. [DOI] [PubMed] [Google Scholar]
- 20.Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, et al. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee I, Kim YO, Park SC, Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 22.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han C, Gronow S, Teshima H, Lapidus A, Nolan M, et al. Complete genome sequence of Treponema succinifaciens type strain (6091) Stand Genomic Sci. 2011;4:361–370. doi: 10.4056/sigs.1984594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almeida A, Nayfach S, Boland M, Strozzi F, Beracochea M, et al. A unified sequence catalogue of over 280000 genomes obtained from the human gut microbiome. bioRxiv. 2019:762682. doi: 10.1101/762682. [DOI] [Google Scholar]
- 25.Pehrsson EC, Tsukayama P, Patel S, Mejía-Bautista M, Sosa-Soto G, et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533:212–216. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, et al. Extensive unexplored human microbiome diversity revealed by oUnexplored Human Microbiome Diversity Revealed by Over 150000 genomes from metagenomes spanning age, geography, and lifeGenomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell. 2019;176:649–662. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cleary B, Brito IL, Huang K, Gevers D, Shea T, et al. Detection of low-abundance bacterial strains in metagenomic datasets by eigengenome partitioning. Nat Biotechnol. 2015;33:1053–1060. doi: 10.1038/nbt.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyss C, Dewhirst FE, Gmür R, Thurnheer T, Xue Y, et al. Treponema parvum sp. nov., a small, glucuronic or galacturonic acid-dependent oral spirochaete from lesions of human periodontitis and acute necrotizing ulcerative gingivitis. Int J Syst Evol Microbiol. 2001;51:955–962. doi: 10.1099/00207713-51-3-955. [DOI] [PubMed] [Google Scholar]
- 29.Nordhoff M, Taras D, Macha M, Tedin K, Busse HJ, et al. Treponema berlinense sp. nov. and Treponema porcinum sp. nov., novel spirochaetes isolated from porcine faeces. Int J Syst Evol Microbiol. 2005;55:1675–1680. doi: 10.1099/ijs.0.63388-0. [DOI] [PubMed] [Google Scholar]
- 30.Evans NJ, Brown JM, Demirkan I, Murray RD, Birtles RJ, et al. Treponema pedis sp. nov., a spirochaete isolated from bovine digital dermatitis lesions. Int J Syst Evol Microbiol. 2009;59:987–991. doi: 10.1099/ijs.0.002287-0. [DOI] [PubMed] [Google Scholar]
- 31.Schrank K, Choi BK, Grund S, Moter A, Heuner K, et al. Treponema brennaborense sp. nov., a novel spirochaete isolated from a dairy cow suffering from digital dermatitis. Int J Syst Bacteriol. 1999;49 Pt 1:43–50. doi: 10.1099/00207713-49-1-43. [DOI] [PubMed] [Google Scholar]
- 32.Wyss C, Choi BK, Schupbach P, Guggenheim B, Gobel UB. Treponema maltophilum sp. nov., a small oral spirochete isolated from human periodontal lesions. Int J Syst Bacteriol. 1996;46:745–752. doi: 10.1099/00207713-46-3-745. [DOI] [PubMed] [Google Scholar]
- 33.Wyss C, Choi BK, Schüpbach P, Guggenheim B, Göbel UB. Treponema amylovorum sp. nov., a saccharolytic spirochete of medium size isolated from an advanced human periodontal lesion. Int J Syst Bacteriol. 1997;47:842–845. doi: 10.1099/00207713-47-3-842. [DOI] [PubMed] [Google Scholar]
- 34.Wyss C, Moter A, Choi B-K, Dewhirst FE, Xue Y, et al. Treponema putidum sp. nov., a medium-sized proteolytic spirochaete isolated from lesions of human periodontitis and acute necrotizing ulcerative gingivitis. Int J Syst Evol Microbiol. 2004;54:1117–1122. doi: 10.1099/ijs.0.02806-0. [DOI] [PubMed] [Google Scholar]
- 35.Wyss C, Choi BK, Schüpbach P, Moter A, Guggenheim B, et al. Treponema lecithinolyticum sp. nov., a small saccharolytic spirochaete with phospholipase A and C activities associated with periodontal diseases. Int J Syst Bacteriol. 1999;49 Pt 4:1329–1339. doi: 10.1099/00207713-49-4-1329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.