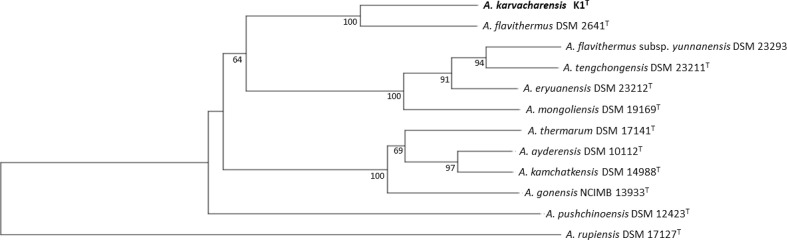
Fig. 2.
Phylogenomic tree of A. karvacharensis strain K1T and related Anoxybacillus type strains. The tree was inferred with FastME 2.1.6.1 [34] from genome blast distance phylogeny (GBDP) distances calculated from genome sequences using the TYGS server (https://tygs.dsmz.de) [35]. The branch lengths are scaled in terms of GBDP distance formula d5. The numbers at branches are GBDP pseudo-bootstrap support values ≥64 % from 100 replications with an average branch support of 97.7 %. The tree was rooted at the midpoint [36]. Genome sequence accession numbers are as follows: A. flavithermus , CP020815.1; A. flavithermus subsp. yunnanensis , GCA_000753835; A. tengchongensis , JACHES000000000.1; A. mongoliensis , JACHEQ000000000.1; A. pushchinoensis , NZ_FOJQ00000000; A. thermarum , JXTH00000000.1; A. ayderensis , JXTG00000000.1; A. kamchatkensis , JACDUV000000000.1, A. eryuanensis , Gp0456347; A. gonensis , CP012152.1; A. rupiensis , Gp0401003.