Abstract
Infliximab is commonly used in inflammatory bowel disease (IBD), however, differences in clinical response among patients are common. Several studies have considered the possibility that these differences are caused by genetic variability even if no unique marker has been yet identified in pediatric patients. We evaluated the impact of two candidate single‐nucleotide polymorphisms (SNPs) rs396991 in FCGR3A and rs1800629 in TNFα genes on infliximab response in an Italian cohort of 76 pediatric patients with IBD. Results showed that patients with the variant FCGR3A allele had a reduced clinical response at the end of induction (p value = 0.004), at 22 weeks (p value = 0.001), and at 52 weeks of treatment (p value = 0.01). A significant association between the FCGR3A variant and median infliximab levels measured during maintenance therapy was also observed: patients with wild type genotype had higher infliximab levels compared to patient with variant allele. Furthermore, patients with the variant allele had a higher probability to produce antidrug antibodies (ADAs). No association was found among the TNFα SNP, clinical response, and infliximab levels. This study addressed for the first time in pediatric patients with IBD, the association of FCGR3A SNP, infliximab response, and ADA production.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
The role of pharmacogenetic factors to predict therapeutic effects in inflammatory bowel disease (IBD) has been largely demonstrated. Anti‐TNFα drugs and ADA concentrations are strongly associated with disease remission, however, differences in response are frequent and often unpredictable. The identification of pharmacogenetic markers may lead to the identification of strategies to reduce treatment failure or loss of response, especially in pediatric patients where the data are lacking.
WHAT QUESTION DID THIS STUDY ADDRESS?
This study addresses for the first time in pediatric patients with IBD, an association of FCGR3A SNP, infliximab response, and ADAs.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
Although data on the importance of some genetic variants in the response to anti‐TNFα drugs in adult patients with IBD are numerous, in the pediatric population they are substantially lacking. This study supports the utility of genotyping FCGR3A gene to predict infliximab response even in this pediatric population.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Future pretreatment genetic testing in genes involved in ADA production, such as FCGR3A, could be evaluated in pediatric patients with IBD treated with anti‐TNFα agents.
INTRODUCTION
Crohn’s disease (CD) and ulcerative colitis (UC), the two major subtypes of inflammatory bowel disease (IBD), are chronic and currently incurable diseases that in 20%–30% of cases occur under the age of 20 years. 1 Chronic gastrointestinal inflammation results in significant morbidity, impaired quality of life, and raised health care utilization and costs. 2 , 3 Pharmacological therapy of IBD, and in particular the use of antibodies against tumor necrosis factor alpha (TNFα), such as infliximab, has improved outcomes. Indeed, these drugs have been shown to induce and maintain disease remission, improving patients’ quality of life and reducing the need for surgery. 4 Several studies have demonstrated the efficacy of these drugs in pediatric patients with IBD 5 , 6 ; however, a significant interindividual variability occurs: 10%–20% of patients lose response during the induction phase (primary failure) and up to 45% have a secondary loss of response. 7 , 8 In addition, in some patients, antidrug antibodies (ADAs) are produced with potential risk of immunogenicity and loss of response. Therefore, anti‐TNFα therapies need optimization through the implementation of therapeutic drug monitoring. 9 , 10 Increasing evidences suggest that these differences are influenced by genetic variability and several studies considering therapeutic response and drug efficacy have been performed, evaluating single nucleotide polymorphisms (SNPs) in genes involved in immune processes and inflammation. 11 , 12
The Fcγ receptor type IIIA (FCGR3A) belongs to a group of surface glycoproteins able to bind monomeric and aggregated immunoglobulin (Ig)G and involved in the removal of antigen‐antibody complexes from the circulation. Infliximab may be removed by opsonization via reticuloendothelial system (RES) following binding of the Fc part of the antibody to FCGR expressed on the RES. The low clearance is due to the low binding affinity of FCGR3A receptor, which maintains the drug into the circulation. 13 , 14 This receptor is expressed on macrophages, natural killer (NK) cells, and monocytes, and mediates antibody‐dependent responses, such as antibody‐dependent cellular cytotoxicity (ADCC) and phagocytosis. 13 , 15 The SNP rs396991 is a nonsynonymous polymorphism in the FCGR3A gene (559A>C, F158V). rs396991 A encodes for a protein with phenylalanine in position 158 and is identified as F allele, that binds with low affinity IgG1 and IgG3, whereas the rs396991 C encodes the variant valine (V) that has a high‐binding affinity for IgG1 and IgG3. 16 Ternant and colleagues were the first to evaluate the influence of FCGR3A SNP on the pharmacokinetics of infliximab in patients with CD, suggesting a possible role of this SNP in drug response. 17
The multifunctional proinflammatory cytokine TNFα has been proposed as a central player in inflammatory cell activation and recruitment and is suggested to play a critical role in the development of IBD. 18 , 19 Many variants in the untranslated region of the human TNFα gene have been described in adult patients affected by IBD and seem to alter the transcriptional process, increasing TNFα production. 20 , 21 In this context, the genetic variant rs1800629 (−308 G>A) has previously been extensively studied and has been associated with susceptibility to a range of autoimmune disorders, among which IBD, and with the response to anti‐TNFα agents in adult patients. 22 , 23 , 24 Although the association between FCGR3A SNP and anti‐TNFα response in adult patients with IBD has been evaluated, 12 , 25 , 26 , 27 no unique marker has been yet identified in pediatric patients.
Therefore, in the present study, our research focused on the impact of FCGR3A rs396991 (559 A>C) and TNFα rs1800629 (−308 G>A) SNPs on infliximab response, and the association between these genetic variants and infliximab concentrations and ADA production in pediatric patients with IBD.
METHODS
Study population and clinical response
Seventy‐six pediatric patients with IBD were enrolled at the Gastroenterology department of the Pediatric Clinic of IRCCS Burlo Garofolo in Trieste and the Pediatric Unit of Ca’ Foncello Hospital in Treviso. The inclusion criteria were age between 7 and 18 years, previous diagnosis of IBD, and treatment with infliximab. Infliximab was started as first‐line therapy or in case of treatment failure or intolerance to first‐line therapies. The therapeutic protocol included an induction phase with intravenous administration of infliximab 5 mg/kg at weeks 0, 2, 6, and then every 8 weeks during the maintenance phase. Concomitant treatment with glucocorticoids, azathioprine, or methotrexate was permitted. In most cases, infliximab and immunosuppressants were started concomitantly. Only in very few cases, immunosuppressants were added after the development of ADAs or low infliximab levels. The exclusion criteria were patients with an ileostomy or colostomy, disease needing surgery, or contemporary presence of other noncontrolled pathologies. Serum samples for infliximab and ADA measurements by enzyme‐linked immunosorbent assays (ELISA), were taken at the appropriate clinical visit both during the induction and maintenance phases. The concentration of infliximab was measured at trough, before the next scheduled administration. The correlation between infliximab level and the different genotypes was assessed during the maintenance phase (16–52 weeks) in a subcohort of 27 pediatric patients with IBD for which both the genotyping and infliximab concentration were available. ADAs were measured when infliximab plasma levels were less than or equal to 1.5 µg/ml. Patients with ADA levels higher than 10 units per milliliter (AU/ml) were considered positive.
For each patient, clinical efficacy was assessed, using Pediatric Crohn’s Disease Activity Index (PCDAI) and Pediatric Ulcerative Colitis Activity Index (PUCAI) for patients with CD and UC, respectively, at the end of induction and during maintenance phase (at 22 and 52 weeks). Disease was considered in remission if indexes were less than or equal to 10.
Ethical consideration
Local ethical committee approval for the study (protocol CEUR‐2018‐OS‐113‐BURLO, number 0035325/P/GEN/EGAS) was provided and appropriate informed consent was obtained from patients and/or their parents or guardians.
DNA extraction and pharmacogenetic analysis
Total gDNA was isolated from patients’ peripheral blood using a commercial kit (Sigma‐Aldrich) according to the manufacturer’s protocol. The DNA concentration and purity were calculated by NanoDrop instrument (NanoDrop 2000, EuroClone). TaqMan SNP genotyping assays (Applied Biosystems), polymerase chain reaction (PCR)‐restriction fragment length polymorphism (RFLP) and/or droplet digital‐PCR (ddPCR) were used to characterize the SNPs of interest (FCGR3A rs396991 and TNFα rs1800629).
PCR SNP analysis and validation
The FCGR3A rs396991 was genotyped with TaqMan Designed SNP Genotyping Assays (C__25815666_10; Thermo Fisher) using the CFX96 real‐time system‐C1000 Thermal Cycler (Bio‐Rad Laboratories). The thermal cycling conditions for TaqMan assays were as follows: 5 min at 95°C followed by 50 cycles at 95°C for 15 s and 60°C for 60 s. Through the software unit (Sequence Detection System) the three different populations’ genotypes (wt, het, and var) were discriminated.
The analysis of the TNFα rs1800629 promoter polymorphisms was determined by PCR‐RFLP. 28
Briefly, the TNFα rs1800629 PCR products obtained by amplification of gDNA (forward and reverse primer sequence: 5′‐GAGCAAAGGTTTTGAGCCCAT‐3′ and 5′‐GGGACACACAAGCATCAAG‐3′) were digested with NcoI (New England Biolabs) and run on a 3.5% SYBR Safe‐stained agarose gel. To confirm the data obtained with PCR‐RFLP, the analysis of this polymorphism was determined also by ddPCR Bio‐Rad’s QX200 system (Bio‐Rad Laboratory). PCR was performed in a Veriti PCR instrument (Thermo Fisher) according to manufacturer's instructions. After thermal cycling, plates were read in the QX200 Droplet Reader. QuantaSoft software was used for analysis (Bio‐Rad Laboratory).
Statistical analysis
Statistical analyses were performed using the software R (version 3.4.2). The association between clinical response and ADA production was evaluated in a univariate analysis by generalized linear models of the binomial family (logistic regression), using clinical response or ADA production as the dependent variable and the two candidate genetic variants, clinical (type of disease) demographic (age and gender), characteristics of population, and concomitant treatment as the independent variables; using the model that considered 0 versus 1 versus 2 alleles. The association between infliximab concentrations and the two candidate genetic variants was assessed by the nonparametric Kruskal‐Wallis test. Multivariate analysis was done by a logistic regression model, which was built selecting dependent variables that had a significant association in the univariate analysis with a p value threshold of 0.05. The association of risk factors with time to produce ADA was assessed by Kaplan‐Meier survival curve a Cox proportional hazards regression models. The standard significance threshold of 0.05 was applied to p values.
RESULTS
Clinical remission
Seventy‐six children in treatment with infliximab were enrolled by the Gastroenterology Unit of IRCCS Burlo Garofolo Hospital in Trieste and the Pediatric Unit of Ca’ Foncello Hospital in Treviso. The demographic characteristics of the population are summarized in Table 1.
TABLE 1.
Demographic and clinical characteristics of the study population
| Patients (n) | 76 |
| Age in years (median, IQR) | 14.7, 12.3–16.3 |
| Males (n, %) | 39 (47.4) |
| CD (n, %) | 50 (65.8) |
| UC (n, %) | 26 (34.2) |
| Concomitant treatment (n) | 39 |
| Glucocorticoid (n, %) | 5 (12.8) |
| Azathioprine (n, %) | 29 (74.4) |
| Azathioprine + glucocorticoid (n, %) | 4 (10.2) |
| Methotrexate (n, %) | 1 (2.6) |
| Primary failure (n, %) | 24 (31.6) |
| Secondary failure: | |
| At 22 weeks a (n, %) | 31 (45.6) |
| At 52 weeks a (n, %) | 30 (45.4) |
| ADA production (n, %) | 13 (56.5) |
Abbreviations: ADA, anti‐drug antibodies; CD, Crohn’s disease; IQR, interquartile range; n, number; UC, ulcerative colitis.
At 22 and 52 weeks of therapy, 8 and 10 patients were lost to follow‐up, respectively.
Considering demographic and clinical variables, neither gender nor age nor IBD type were significantly associated with infliximab clinical response at the end of induction (p = 0.18, p = 0.10, and p = 0.80, respectively), at 22 weeks (p = 0.12, p = 0.96, and p = 0.47, respectively) and with ADA production (p = 0.10, p = 0.17, and p = 0.81, respectively) after univariate logistic regression analysis. Considering clinical response at 52 weeks, a significant association with gender was found, with an increased therapy failure in girls compared to males (p = 0.03), whereas no association was found with age and IBD type (p = 0.40 and p = 0.21, respectively). Evaluating the effect of concomitant treatment with immunosuppressants on infliximab response, no association was found with the response at the end of induction (p = 0.43), at 22 weeks (p = 0.82), at 52 weeks (p = 0.63), and with ADA production (p = 0.40).
Association between FCGR3A and TNFα candidate variants and clinical response
The genotype distribution of the FCGR3A and TNFα SNPs in the study population is summarized in Table 2. Genotype frequencies were in Hardy–Weinberg equilibrium (HWE; p > 0.05).
TABLE 2.
FCGR3A and TNFα SNPs and genotype distribution of pediatric patients with IBD
| Gene | SNP |
Position (Assembly hg38) |
IBD pediatric patients (N = 76) | |||
|---|---|---|---|---|---|---|
| Wt (%) | Het (%) | Var (%) | HWE | |||
| FCGR3A | rs396991 |
chr1:161544752; 559 A>G (V158F) |
33 (43.4) | 29 (38.2) | 14 (18.4) | 0.1 |
| TNFα | rs1800629 |
chr6:31575254; −308 G>A |
59 (77.6) | 15 (19.8) | 2 (2.6) | 0.3 |
A significant association between the FCGR3A SNP and clinical response at the end of induction (p = 0.004, odds ratio [OR] FF vs. VF genotype = 6.76, confidence interval [CI] = 1.89–24.19; OR FF vs. VV genotype = 5.43, CI = 1.22–24.07), at 22 weeks (p = 0.001, OR FF vs. VF genotype = 8.89, CI = 2.52–31.41; OR FF vs. VV genotype = 4.39, CI = 1.05–18.35), and at 52 weeks (p = 0.01, OR FF vs. VF genotype = 6.58, CI = 1.91–23.17; OR FF vs. VV genotype = 2.5, CI = 0.61–10.11) was observed (Figure 1 and Supplementary Table S1). No significant association was found between the TNFα SNP and clinical response at the end of induction (p = 0.84, OR GG vs. AG genotype = 1.13, CI = 0.34–3.81; OR GG vs. AA genotype = 2.27, CI = 0.13–38.50), at 22 weeks (p = 0.72, OR GG vs. AG genotype = 1.75, CI = 0.44–6.89; OR GG vs. AA genotype = 1.16, CI = 0.07–19.67), and at 52 weeks (p = 0.68, OR GG vs. AG genotype = 1.94, CI = 0.42–8.99; OR GG vs. AA genotype = 1.19, CI = 0.07–19.69; Figure 2). No association was found between the use of concomitant immunosuppressants treatment and the different genotypes (p > 0.05) and the same result was also obtained considering the demographical variables (age and gender) and disease type.
FIGURE 1.
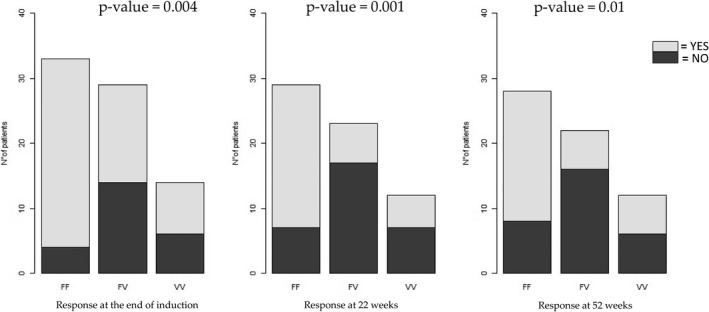
Association between FCGR3A SNP rs396991 and clinical response to infliximab in 76 pediatric patients with inflammatory bowel disease at the end of induction (14 weeks of therapy) or during maintenance (at 22 and 52 weeks of therapy). Logistic regression was used for statistical analysis
FIGURE 2.
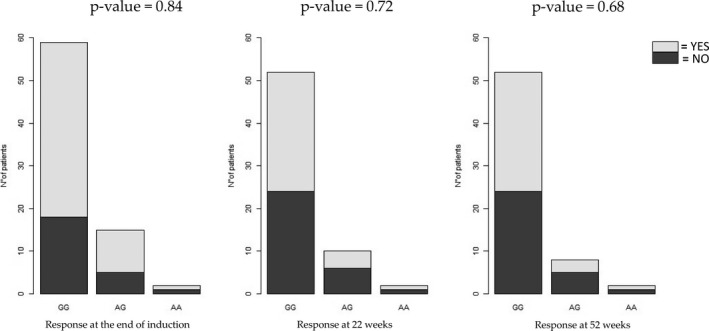
Lack of association between TNFα SNP rs1800629 and clinical response to infliximab in 76 pediatric patients with inflammatory bowel disease at the end of induction (14 weeks of therapy) or during maintenance (at 22 and 52 weeks of therapy). Logistic regression was used for statistical analysis
Association between serum infliximab concentrations, ADA production and FCGR3A and TNFα candidate variants
The association between infliximab pharmacokinetics and FCGR3A and TNFα polymorphisms was evaluated in a subgroup of patients (n = 27), based on the availability of patient’s sera for infliximab quantification. The data show a significant association between the FCGR3A variant and median infliximab levels measured during maintenance phase: patients with the FF genotype had higher infliximab levels compared to patients with the variant allele V (5.40 μg/ml, interquartile range [IQR]: 5.40; vs. 1.31 μg/ml, IQR: 1.28; vs. 4.20 μg/ml, IQR: 0.21; for FF, VF and VV respectively; p = 0.02). No significant association was found between serum infliximab concentration and the TNFα variant (3.45 μg/ml, IQR: 2.50; vs. 4.60 μg/ml, IQR: 3.42; vs. 2.85 μg/ml, IQR: 0; for GG, AG, and AA, respectively; p = 0.84; Figure 3).
FIGURE 3.
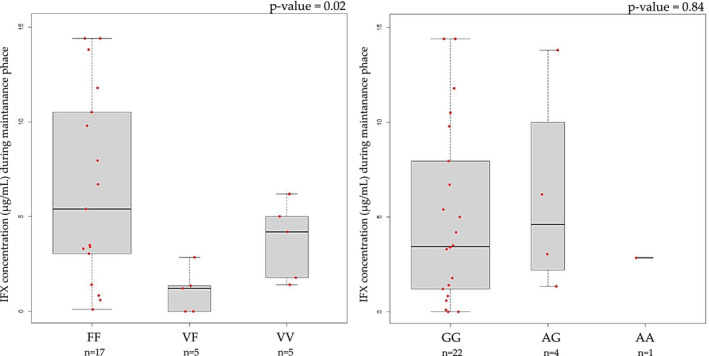
Boxplot comparing serum infliximab (IFX) concentration and candidate variants. The bold horizontal line represents the median value. Statistical significance was assessed by Kruskal Wallis test
A significant association was found between FCGR3A SNP and ADA production (p = 0.01). In particular, patients with the variant allele V had a higher probability to produce ADA (OR FF vs. VF genotype = 11.90, CI = 1.85–76.53; OR FF vs. VV genotype = 5.10, CI = 0.65–39.54). No association was found between TNFα SNP and ADA production (p = 0.63, OR GG vs. AG genotype = 1.40, CI = 0.27–7.15; OR GG vs. AA genotype = 0.75, CI = 0.03–20.26; Figure 4).
FIGURE 4.
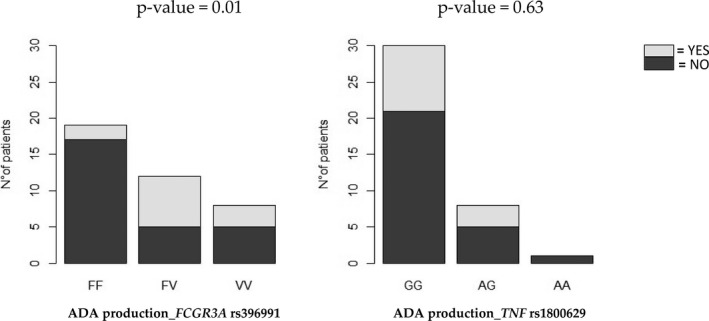
Association between production of antidrug antibodies (ADAs) during maintenance therapy with infliximab according to FCGR3A and TNFα genotypes. Logistic regression was used for statistical analysis
The analysis of proportional risk over the 52‐week follow‐up reveals that patients carrying the FCGR3A variant V allele had a significantly increased risk of ADA production compared to homozygous wild‐type patients (hazard ratio = 7.3, CI = 1.59–33.6, p = 0.01; Figure 5). No significant difference in proportional risk of ADA production could be observed according to the TNFα genotype (Supplementary Figure S1).
FIGURE 5.
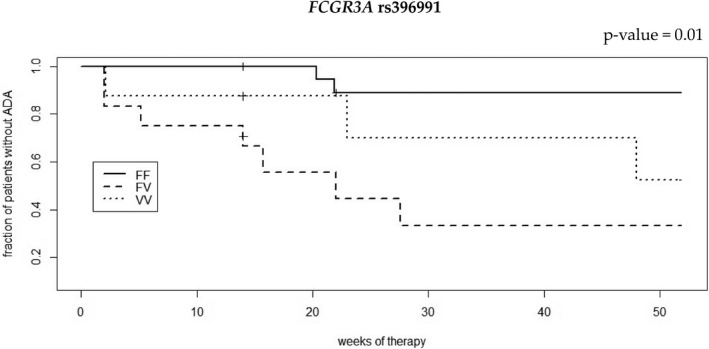
Survival curves of infliximab anti‐drug antibodies (ADAs) production according to FCGR3A rs396991 genotype from a Cox proportional hazard model. Vertical dashes indicate patients (n = 6) that have interrupted the follow‐up before week 52
Multivariate analysis
Multivariate analysis was done by logistic regression, using therapeutic response (at week 52) as the dependent variable and all covariates (gender and FCGR3A SNP) significantly associated in the univariate analysis. Multivariate analysis confirmed that FCGR3A SNP was associated with clinical response at 52 weeks of treatment with infliximab (adjusted p = 0.01), but not gender (adjusted p = 0.08). Analysis evaluating the association of clinical response with infliximab concentrations and FCGR3A genotypes as independent variables, was done also in the subcohort of 27 patients that have both the genotyping and infliximab concentration information available. These analyses support the effect of the median infliximab concentration on drug response (p < 0.05); however, we could not verify an effect of the genotype (p > 0.05) likely because of the limited size of this subgroup.
DISCUSSION
This study identified for the first time the role of FCGR3A rs396991 polymorphism as useful genetic biomarker of infliximab response in a retrospective cohort of pediatric patients with IBD. The functional polymorphism in FCGR3A gene rs396991 (F158V) has been reported to influence human IgG1 binding and ADCC activity. 29 A significant association between the FCGR3A SNP and loss of response at the end of induction and during maintenance phase (at 22 and 52 weeks) was found in our pediatric cohort. Several reports have investigated the association between FCGR3A SNP and anti‐TNFα response in adult patients with chronic inflammatory disorders, 12 , 25 , 26 , 27 even if results are not always univocal, maybe because clinical response is influenced by several factors, such as concomitant treatments. 30 To our knowledge, no study has evaluated yet this association in pediatric patients with IBD. Ternant and colleagues were the first to demonstrate the influence of this SNP on the pharmacokinetics of infliximab in patients with CD, suggesting that the higher binding affinity of the FCGR to IgG in patients carrying the V variant alleles could induce a higher infliximab elimination rate assuming a possible increase in NK cells recruitment and ADCC activity and resulting in a underexposure to infliximab and relapse. 17 These considerations were supported by Moroi and colleagues that demonstrated that NK cells isolated from FCGR3A V patients had a higher infliximab binding affinity than those from FF subjects and a higher ADCC activity in vitro that could influence biological responses to infliximab in patients with CD. 15 Considering the potential mechanism by which the FCGR3A variant influences infliximab response, in our study, in a subcohort of patients for which infliximab concentration was also available, pediatric patients with variant V allele showed lower levels of infliximab compared to FF patients. The FCGR3A genotypes may have a pharmacodynamic effect in addition to a pharmacokinetic effect on infliximab clearance, however, further studies on larger cohorts of pediatric patients are needed to shed light on this potential effect.
The present trend for which the heterozygous group has the lowest infliximab concentrations as compared to other genotypes, seems not consistent with a mechanistic conclusion according to which the variant increases the affinity of the receptor for Fc fragment. We hypothesize that the effect of the FCGR3A variant may be dominant, as supported by our results on the association between clinical response, evaluated at the end of induction and at 22 weeks of therapy, and the genotypes, and also by other published results. 15
The production of antibodies against anti‐TNFα agents is an important cause of loss of response in patients with IBD. 31 ADA production is associated with worse treatment response, due to a lower bioavailability caused by higher anti‐TNFα clearance. Indeed, the generation of ADAs facilitates the opsonization and phagocytosis of anti‐TNFα agents. Our results show that pediatric patients with the variant allele V have a higher probability to produce ADAs, in line with previous studies in adult patients with IBD, treated with infliximab and adalimumab. 32 The proinflammatory TNFα cytokine plays an important role in the pathogenesis of IBD and it is possible that different levels of this cytokine may affect the response to therapy. Many variants in the untranslated region of the human TNFα gene have also been described and seem to alter the transcriptional process, increasing TNFα production. 31 Several studies concerning the association of TNFα rs1800629 polymorphism and response to anti‐TNFα agents in inflammatory disorders have been performed. 23 , 28 , 33 A meta‐analysis of nine studies concludes that rs1800629 A allele carriers respond less to TNFα inhibitors when used in the treatment of rheumatoid arthritis, probably due to higher TNFα levels. 24 Similar results were demonstrated in adult patients with IBD, 26 , 33 , 34 whereas no data are available for pediatric patients. Contrary to what was demonstrated in adults, our study could not find an association between TNFα SNP and anti‐TNFα response. A meta‐analysis on the effect of the TNFα SNP on infliximab response in adults underlined the necessity of a cohort size of 200 patients with IBD to obtain a significant association between the TNFα SNP and clinical response. 35 For this reason, the lack of effect of TNFα SNP observed may be due to the limited statistical power of our study. Further analysis will be performed in a larger cohort of pediatric patients with IBD, to assess the possible role of TNFα SNP in infliximab response. Similar considerations could be valid also for other covariates, such as concomitant immunosuppressants. 36
In conclusion, although the present study has some limitations regarding the small sample size and its retrospective nature, this is the first time that new information about the role of FCGR3A SNP rs396991 in pediatric patients with IBD are provided: this SNP seems to affect infliximab response and influence ADA’s production. Validation in a larger cohort is required and, if these data are confirmed, future pretreatment genetic testing in genes involved in ADA production, such as FCGR3A, could be suggested in order to achieve a personalized therapy in pediatric patients with IBD.
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
D.C., M.L., G.S., and R.F. wrote the manuscript. G.S. and G.D. designed the research. D.C., M.F., A.C., M.B., and S.M. performed the research. D.C., M.L., and G.S. analyzed the data.
Supporting information
Fig S1
Table S1
Curci D, Lucafò M, Cifù A, et al. Pharmacogenetic variants of infliximab response in young patients with inflammatory bowel disease. Clin Transl Sci. 2021;14:2184–2192. 10.1111/cts.13075
Funding information
This study was supported by the Institute for Maternal and Child Health “Burlo Garofolo,” Trieste, Italy (grant number RC 1/17).
REFERENCES
- 1. Castro M, Papadatou B, Baldassare M, et al. Inflammatory bowel disease in children and adolescents in Italy: data from the pediatric national IBD register (1996–2003). Inflamm Bowel Dis. 2008;14:1246‐1252. [DOI] [PubMed] [Google Scholar]
- 2. Burisch J, Vardi H, Schwartz D, et al. Health‐care costs of inflammatory bowel disease in a pan‐European, community‐based, inception cohort during 5 years of follow‐up: a population‐based study. Lancet Gastroenterol Hepatol. 2020;5(5):454‐464. [DOI] [PubMed] [Google Scholar]
- 3. Burisch J, Jess T, Martinato M, Lakatos PL. The burden of inflammatory bowel disease in Europe. J Crohns Colitis. 2013;7:322‐337. [DOI] [PubMed] [Google Scholar]
- 4. Papamichael K, Lin S, Moore M, Papaioannou G, Sattler L, Cheifetz AS. Infliximab in inflammatory bowel disease. Ther Adv Chronic Dis. 2019;10:2040622319838443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hyams J, Crandall W, Kugathasan S, et al. Induction and maintenance infliximab therapy for the treatment of moderate‐to‐severe Crohn's disease in children. Gastroenterology. 2007;132:863‐873. [DOI] [PubMed] [Google Scholar]
- 6. Faubion WA, Dubinsky M, Ruemmele FM, et al. Long‐term efficacy and safety of adalimumab in pediatric patients with Crohn's disease. Inflamm Bowel Dis. 2017;23:453‐460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Naviglio S, Lacorte D, Lucafò M, et al. Causes of treatment failure in children with inflammatory bowel disease treated with infliximab: a pharmacokinetic study. J Pediatr Gastroenterol Nutr. 2019;68:37‐44. [DOI] [PubMed] [Google Scholar]
- 8. Jossen J, Dubinsky M. Therapeutic drug monitoring in inflammatory bowel disease. Curr Opin Pediatr. 2016;28:620‐625. [DOI] [PubMed] [Google Scholar]
- 9. Franca R, Curci D, Lucafò M, Decorti G, Stocco G. Therapeutic drug monitoring to improve outcome of anti‐TNF drugs in pediatric inflammatory bowel disease. Expert Opin Drug Metab Toxicol. 2019;15:527‐539. [DOI] [PubMed] [Google Scholar]
- 10. Curci D, Lucafò M, Cifù A, et al. Determination of serum infliximab concentration by point‐of‐care devices in children with inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2019;69:474‐479. [DOI] [PubMed] [Google Scholar]
- 11. Lucafò M, Franca R, Selvestrel D, et al. Pharmacogenetics of treatments for inflammatory bowel disease. Expert Opin Drug Metab Toxicol. 2018;14:1209‐1223. [DOI] [PubMed] [Google Scholar]
- 12. Prieto‐Pérez R, Almoguera B, Cabaleiro T, Hakonarson H, Abad‐Santos F. Association between genetic polymorphisms and response to anti‐TNFs in patients with inflammatory bowel disease. Int J Mol Sci. 2016;17:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mahaweni NM, Olieslagers TI, Rivas IO, et al. A comprehensive overview of FCGR3A gene variability by full‐length gene sequencing including the identification of V158F polymorphism. Sci Rep. 2018;8(1):15983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keizer RJ, Huitema AD, Schellens JH, Beijnen JH. Clinical pharmacokinetics of therapeutic monoclonal antibodies. Clin Pharmacokinet. 2010;8:493‐507. [DOI] [PubMed] [Google Scholar]
- 15. Moroi R, Endo K, Kinouchi Y, et al. FCGR3A‐158 polymorphism influences the biological response to infliximab in Crohn's disease through affecting the ADCC activity. Immunogenetics. 2015;9:545. [DOI] [PubMed] [Google Scholar]
- 16. Koene HR, Kleijer M, Algra J, Roos D, Von dem Borne AE, De Haas M. Fc gammaRIIIa‐158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa‐48L/R/H phenotype. Blood. 1997;1(90):1109‐1114. [PubMed] [Google Scholar]
- 17. Ternant D, Berkane Z, Picon L, et al. Assessment of the influence of inflammation and FCGR3A genotype on infliximab pharmacokinetics and time to relapse in patients with Crohn's disease. Clin Pharmacokinet. 2015;54:551‐562. [DOI] [PubMed] [Google Scholar]
- 18. Leppkes M, Neurath MF. Cytokines in inflammatory bowel diseases ‐ update. Pharmacol Res. 2020;2020:158‐104835. [DOI] [PubMed] [Google Scholar]
- 19. Ruder B, Atreya R, Becker C. Tumour necrosis factor alpha in intestinal homeostasis and gut related diseases. Int J Mol Sci. 2019;16:20‐1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Balding J, Livingstone WJ, Conroy J, et al. Inflammatory bowel disease: the role of inflammatory cytokine gene polymorphisms. Mediators Inflamm. 2004;13:181‐187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bank S, Andersen PS, Burisch J, et al. Associations between functional polymorphisms in the NFκB signaling pathway and response to anti‐TNF treatment in Danish patients with inflammatory bowel disease. Pharmacogenomics J. 2014;14:526‐534. [DOI] [PubMed] [Google Scholar]
- 22. Fragoso JM, Vargas‐Alarcón G, Jiménez‐Morales S, Reyes‐Hernández OD, Ramírez‐Bello J. Tumor necrosis factor alpha (TNF‐α) in autoimmune diseases (AIDs): molecular biology and genetics. Gac Med Mex. 2014;150:334‐344. [PubMed] [Google Scholar]
- 23. López‐Hernández R, Valdés M, Campillo JA, et al. Genetic polymorphisms of tumour necrosis factor alpha (TNF‐α) promoter gene and response to TNF‐α inhibitors in Spanish patients with inflammatory bowel disease. Int J Immunogenet. 2014;41:63‐68. [DOI] [PubMed] [Google Scholar]
- 24. O'Rielly DD, Roslin NM, Beyene J, Pope A, Rahman P. TNF‐alpha‐308 G/A polymorphism and responsiveness to TNF‐alpha blockade therapy in moderate to severe rheumatoid arthritis: a systematic review and meta‐analysis. Pharmacogenomics J. 2009;9:161‐167. [DOI] [PubMed] [Google Scholar]
- 25. Cartron G, Dacheux L, Salles G, et al. Therapeutic activity of humanized anti‐CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 2002;99:754‐758. [DOI] [PubMed] [Google Scholar]
- 26. Louis E, El Ghoul Z, Vermeire S, et al. Association between polymorphism in IgG Fc receptor IIIa coding gene and biological response to infliximab in Crohn's disease. Aliment Pharmacol Ther. 2004;19:511‐519. [DOI] [PubMed] [Google Scholar]
- 27. Cañete JD, Suárez B, Hernández MV, et al. Influence of variants of Fc gamma receptors IIA and IIIA on the American College of Rheumatology and European League against Rheumatism responses to anti‐tumour necrosis factor alpha therapy in rheumatoid arthritis. Ann Rheum Dis. 2009;68:1547‐1552. [DOI] [PubMed] [Google Scholar]
- 28. Fabris M, Quartuccio L, Fabro C, et al. The ‐308 TNFα and the ‐174 IL‐6 promoter polymorphisms associate with effective anti‐TNFα treatment in seronegative spondyloarthritis. Pharmacogenomics J. 2016;16:238‐242. [DOI] [PubMed] [Google Scholar]
- 29. Tutuncu Z, Kavanaugh A, Zvaifler N, Corr M, Deutsch R, Boyle D. Fcgamma receptor type IIIA polymorphisms influence treatment outcomes in patients with inflammatory arthritis treated with tumor necrosis factor alpha‐blocking agents. Arthritis Rheum. 2005;52:2693‐2696. [DOI] [PubMed] [Google Scholar]
- 30. Louis EJ, Watier HE, Schreiber S, et al. Polymorphism in IgG Fc receptor gene FCGR3A and response to infliximab in Crohn's disease: a subanalysis of the ACCENT I study. Pharmacogenet Genomics. 2006;16:911‐914. [DOI] [PubMed] [Google Scholar]
- 31. Cohen RZ, Schoen BT, Kugathasan S, Sauer CG. Management of anti‐drug antibodies to biologic medications in children with inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2019;69:551‐556. [DOI] [PubMed] [Google Scholar]
- 32. Romero‐Cara P, Torres‐Moreno D, Pedregosa J, et al. A FCGR3A polymorphism predicts anti‐drug antibodies in chronic inflammatory bowel disease patients treated with anti‐TNF. Int J Med Sci. 2018;15:10‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Louis E, Vermeire S, Rutgeerts P, et al. A positive response to infliximab in Crohn disease: association with a higher systemic inflammation before treatment but not with ‐308 TNF gene polymorphism. Scand J Gastroenterol. 2002;37:818‐824. [PubMed] [Google Scholar]
- 34. Netz U, Carter JV, Eichenberger MR, et al. Genetic polymorphisms predict response to anti‐tumor necrosis factor treatment in Crohn's disease. World J Gastroenterol. 2017;21:4958‐4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Song GG, Seo YH, Kim JH, Choi SJ, Ji JD, Lee YH. Association between TNF‐α (‐308 A/G, ‐238 A/G, ‐857 C/T) polymorphisms and responsiveness to TNF‐α blockers in spondyloarthropathy, psoriasis and Crohn's disease: a meta‐analysis. Pharmacogenomics. 2015;12:1427‐1437. [DOI] [PubMed] [Google Scholar]
- 36. Colombel JF, Adedokun OJ, Gasink C, et al. Combination therapy with infliximab and azathioprine improves infliximab pharmacokinetic features and efficacy: a post hoc analysis. Clin Gastroenterol Hepatol. 2019;8:1525‐1532. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1


