Fig. 5. Activation of phosphoinositide second messengers and signals.
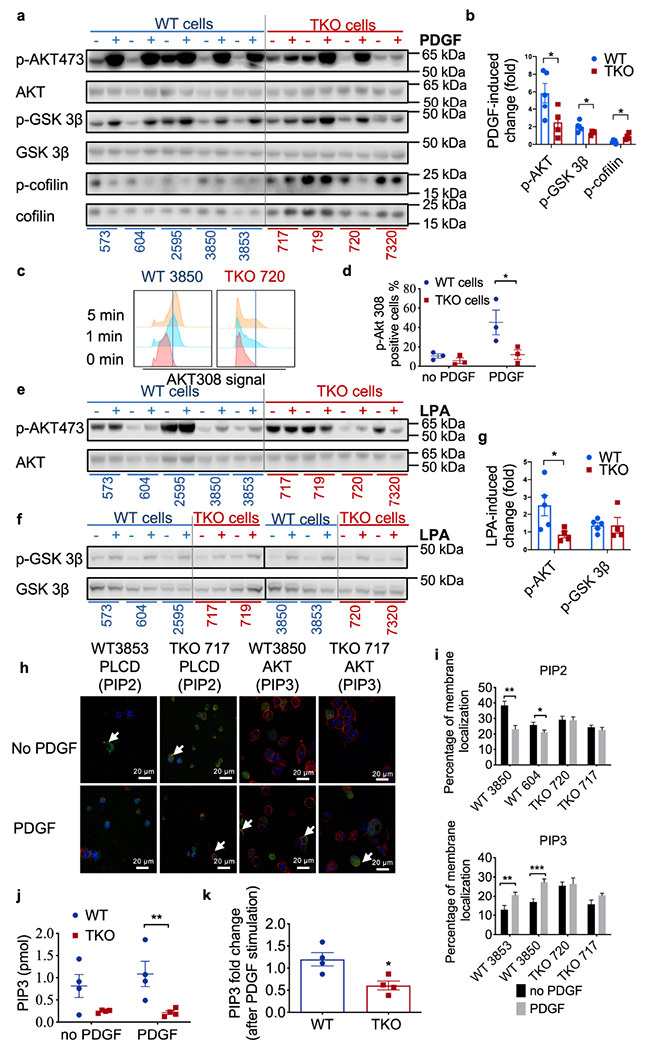
a,b, AKT, GSK-3β and cofilin protein expression in lysates of fibrosarcoma cells cultured with (+) or without (−) PDGF stimulation for 20 min as determined by Western blot (a), and quantification for 5 WT and 4 TKO cell lines (b, p=0.0468 for p-AKT, p=0.047 for p-GSK-3β, p=0.0225 for cofilin). Representative of 3 experiments.
c,d, Phosphorylated AKT308 abundance in WT 3850 and TKO 720 fibrosarcoma cells with (+) or without (−) PDGF stimulation on for 0, 1 and 5 min as determined by Flow cytometry (c) and its quantification for WT 604, 3850, 3853 and TKO 717, 720, 7320 cells (d, p=0.0103). Representative of 4 experiments.
e-g, AKT, GSK-3β and YAP protein expression in lysates of fibrosarcoma cells cultured with (+) or without (−) LPA stimulation for 1 hour as determined by Western blot and its quantification for 5 WT and 4 TKO cells (g, p=0.0463). Representative of 2 experiments.
h,i, Visualization of PLCD-PH-GFP and AKT-PH-GFP by confocal microscopy, with cell membrane stained with WGA and nuclei stained with DAPI in WT and TKO cells. White arrows indicate GFP signals on membrane. Two cell lines were used for each genotype. Cells (n=50) in five randomly selected fields were selected for the quantification of PIP2 and PIP3 signals on membrane (i, p=0.0012 and 0.0434 for PIP2, p=0.0061 and 0.0003 for PIP3). Scale bars = 20 μm. Representative of 4 experiments.
j,k, PIP3 amount in 4 WT (573, 604, 2595, 3850 and 3853) and 4 TKO (717, 719, 720 and 7320) cells with and without PDGF stimulation detected by ELISA (j, p=0.008), and calculated PIP3 fold changes before and after stimulation (k, p=0.0172). Representative of 2 independent experiments. For all graphs, *p < 0.05; **p < 0.01; ***p < 0.001 vs WT; data are presented as mean ± SEM. Statistical significance was determined by two-tailed unpaired t-test (b, i and k) and multiple t test (d, g and j).
