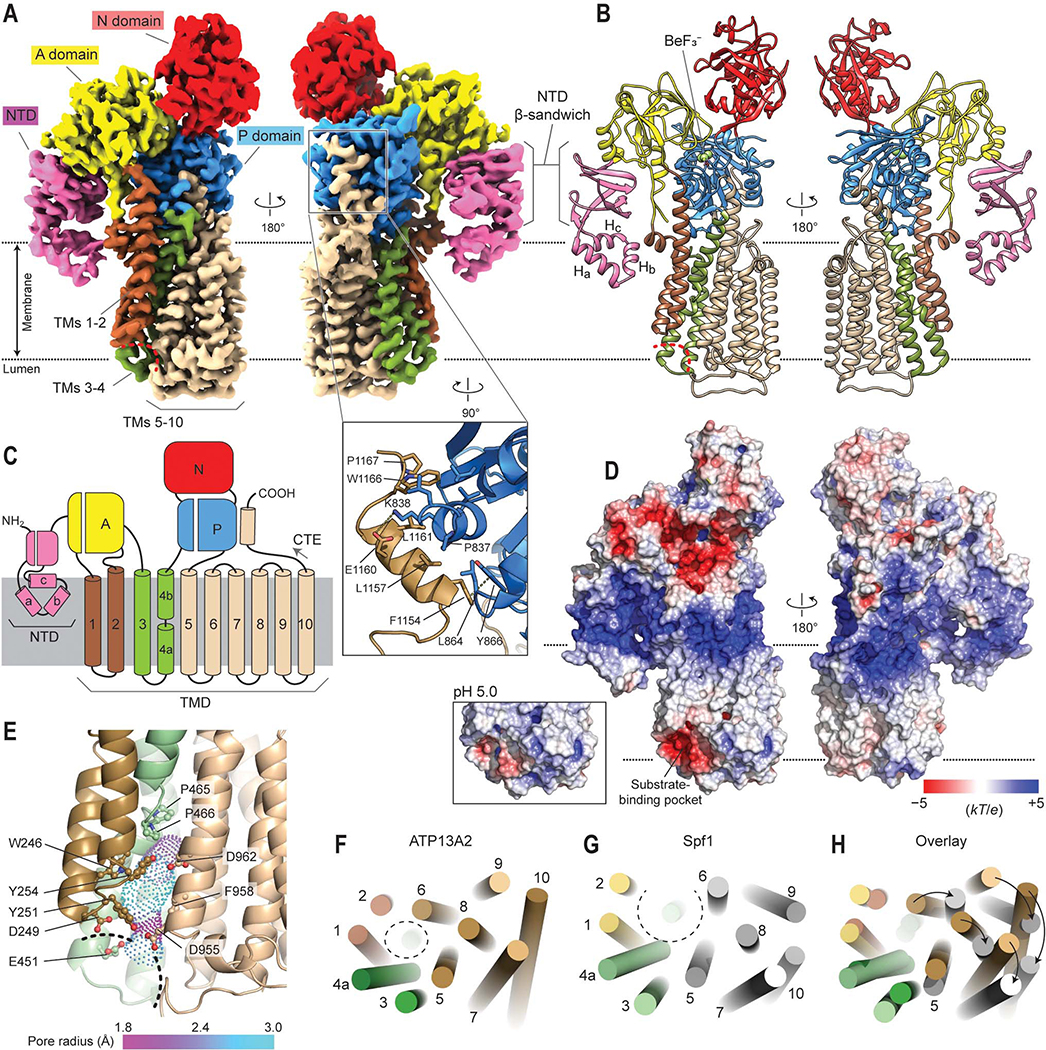
Figure 1. Cryo-EM structure of human ATP13A2 in the E2P state.
(A) Cryo-EM map of human ATP13A2 in the E2·BeF3– form (map 3). Left, a front view; right, a view from the back. Red dashed line, entrance of the substrate-binding pocket. Inset, a zoomed view into the C-terminal extension (CTE) region. Amino acid side chains at the interface between the P domain and CTE are shown as sticks. (B) Atomic model built into the map in A. (C) Domain architecture of ATP13A2. Note that the TM4 helix is unraveled in the middle (between TM4a and TM4b) by a PP(A/V)xP motif conserved among P5B-ATPases. (D) Estimated surface electrostatic potential at pH 7. Inset, electrostatics around the substrate binding pocket estimated at pH 5. (E) View of the substrate-binding pocket in the E2P structure. Dashed line indicates entrance of substrate-binding pocket. Amino acid side chains (balls and sticks) lining the cavity (dots) are shown. Pore radius is indicated by a color gradient. (F–H) Comparison between ATP13A2 and Spf1 structures. TM helices are represented by cylinders. Views are from the lumen. The substrate-binding pockets are indicated by dashed circles.