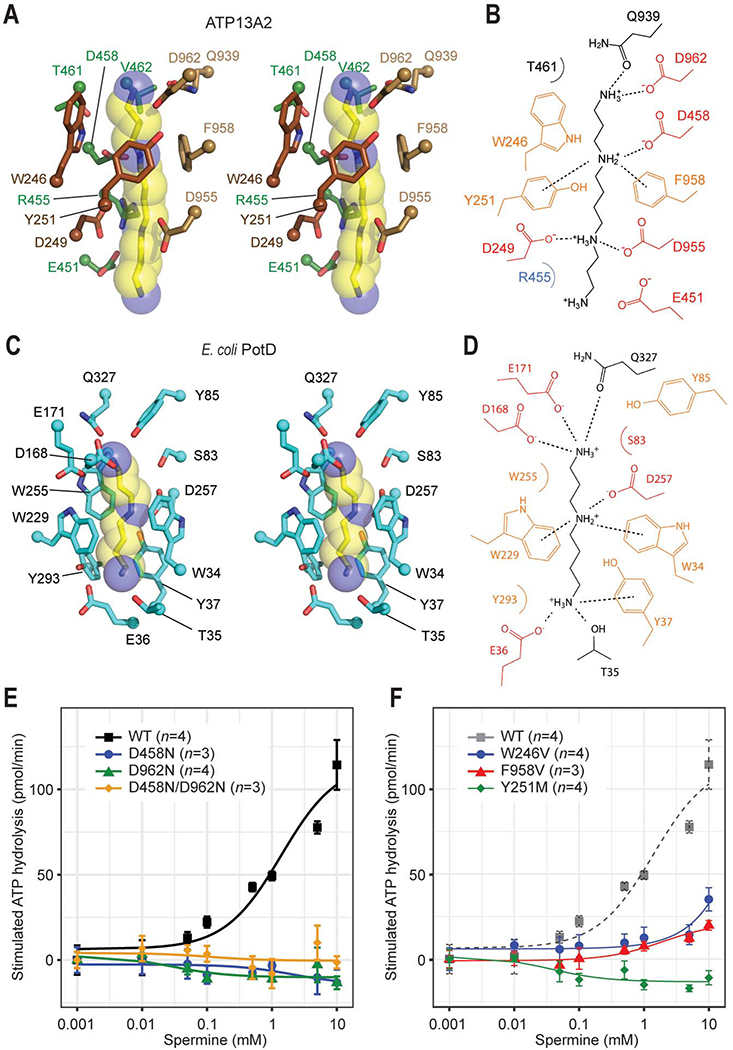
Figure 3. Structural basis of polyamine binding in ATP13A2.
(A) Stereo view of the polyamine-binding site of ATP13A2 in the E2-Pi structure. The modelled spermine molecule is shown as yellow (carbon)/blue (nitrogen) sticks and semitransparent spheres. Side chains of ATP13A2 coordinating the spermine molecule are shown in a stick representation (brown, TMs 1–2; green TM4; tan, TM6). (B) Interactions between ATP13A2 and the polyamine (spermine) molecule. Dashed lines, ionic and cation-π interactions. (C) As in A, but with the spermidine-bound Escherichia coli PotD (PDB: 1POT). (D) As in B, but for the PotD-spermidine structure. (E and F) Spermine-induced ATPase stimulation of microsomes expressing WT ATP13A2 or mutants of polyamine-interacting aspartate (E) or aromatic (F) residues (means and s.e.m.). Lines are fitted dose-response (Michaelis-Menten) curves.