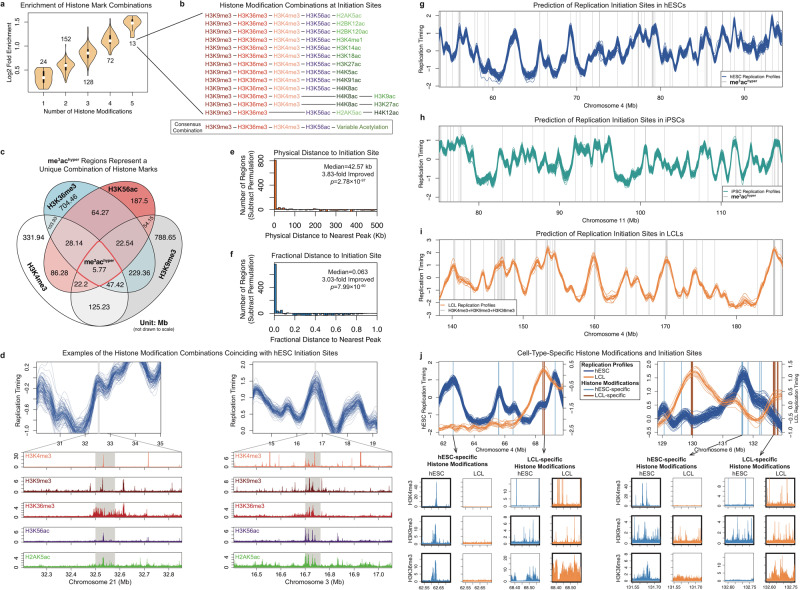
Fig. 3. Histone Modifications Associated with Replication Initiation.
a Iterative identification of histone mark combinations enriched at rtQTLs. Shown are enrichment distributions; the number of combinations in each category is indicated. Fold-enrichment increases gradually and is maximal for five-mark combinations. n indicated in the plot, unit: histone mark combinations. b Histone modification combinations for human replication initiation. The 13 combinations of five histone marks converged to a consensus combination. See Supplementary Table 1 for additional details. c The histone modifications (independent of rtQTLs) represents a rare combination of both active and repressive histone marks. me3achyper regions comprised 0.7–3% of the regions that carry the individual histone marks. d Examples of histone mark combinations (Roadmap Epigenomics imputation)72,73 coinciding with replication timing peaks not identified as rtQTLs. e, f Distribution (after subtraction of permutations) of physical (e) and fractional distances (f) of the me3achyper locations to the nearest replication timing peak. We also indicate the fold-improvement in median distance to the nearest peak compared with random expectation. See Supplementary Fig. 5c, d for further comparison with random. Panels e and f used a one-sided Wilcoxon rank-sum test. Correction for multiple testing was not applicable, as there was only one test for each sub-plot. g Combination of histone marks (gray, me3achyper locations) predict replication initiation sites in hESCs. h, i Histone modification locations (gray vertical lines) correspond to replication timing peaks in iPSCs (h) and LCLs (i). j Cell-type-specific histone modification locations mark cell-type-specific replication initiation sites. At regions with distinct replication timing profiles for hESCs and LCLs, LCL (hESC)-specific replication timing peaks are predicted by LCL (hESC)-specific histone modification locations. Lower panels: initiation sites coincide (thick borders) with all three histone trimethylation marks in the cell type in which they are active, but with one or none of the marks in the cell type in which they are inactive.