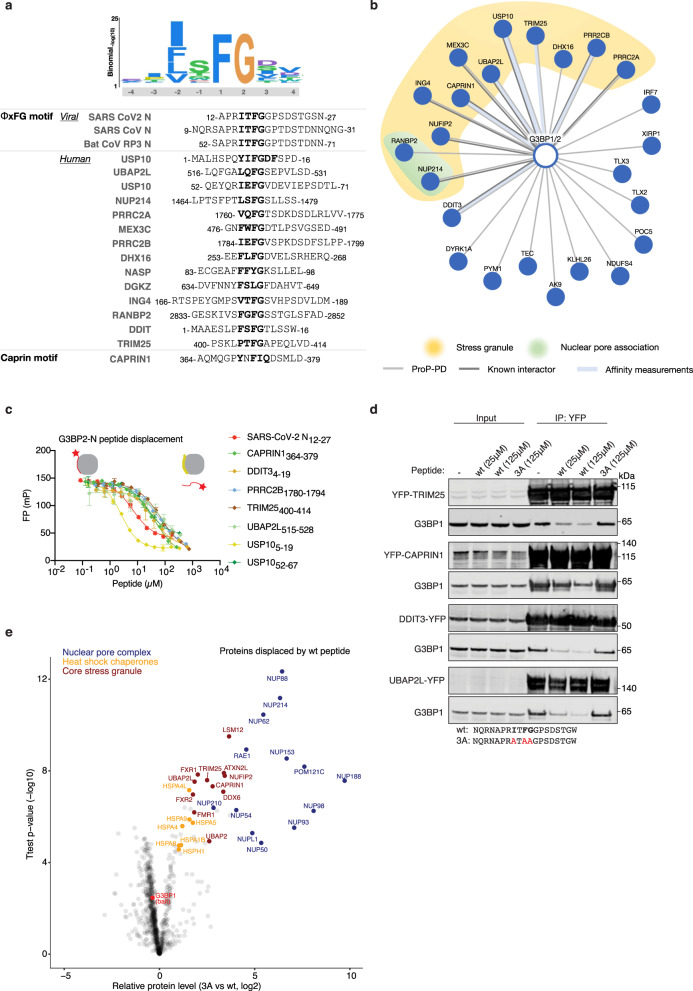
Fig. 4. N competes with cellular proteins for binding to G3BP1/2 through its ΦxFG motif.
a Schematic of the position-specific scoring matrix ΦxFG for G3BP NTF2 domains and sequence alignment of coronavirus peptides and selected human G3BPs ligands found through ProP-PD. b Network of a select set of human SLiM-based interactions of the G3BPs found through ProP-PD. Light grey line indicates interactions validated by affinity measurements (Supplementary Data 3), the weight of the line represents the affinity of the interaction (thick, 1–10 μM; medium, 11–100 μM; thin, 101–500 μM). Dark grey lines indicate protein-protein interactions with additional reported evidence. c The SARS-CoV-2 ΦxFG N peptide competes with the indicated peptides for G3BP2 binding in vitro (n= two biological duplicates each containing three technical replicates). Shown is a representative plot. Error bars expressed as mean ± SD. d The N ΦxFG peptide but not a control peptide competes with TRIM25, CAPRIN1, DDIT2 and UBAP2L for binding to G3BP1 in cells. Shown is a representative blot from three independent experiments. e G3BP1-YFP was affinity purified from HeLa cells in the presence of either a SARS-CoV-2 ΦxFG N wt or N 3 A peptide and analysed by quantitative mass spectrometry. The volcano plot shows the two-sided unpaired T test results for all quantified proteins based on four technical replicates per condition.