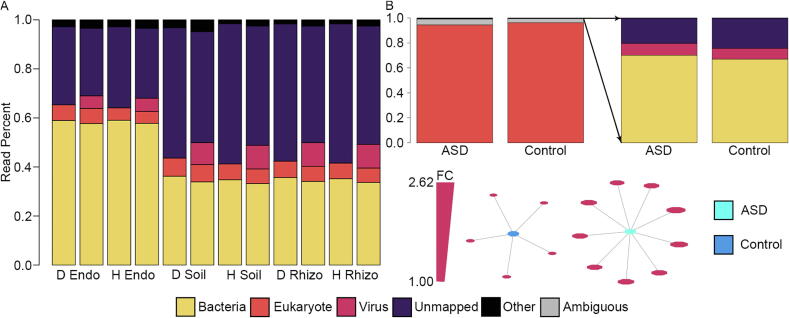
Fig. 2.
Classification of viruses in metagenomic and metatranscriptomic samples. A) Effect of the virus databases on the number of reads mapped in the Populus deltoides (D) and hybrid (H) data. The first bar in each group represents ParaKraken results before the viral databases were included and the second bar represents classification after the inclusion of the viral databases. Metagenome viruses averaged 15% of the mapped reads with a higher percent mapping in the rhizosphere (Rhizo) and soil (Soil) relative to the endosphere (Endo), suggesting that viruses make up a substantial portion of the microbiome. B) Differences in viruses between ASD brains and control brains. Unsurprisingly, eukaryotes make up the vast majority of reads in human samples. However, we were able to identify sequences associated with viruses, eight of which were significantly higher in ASD versus controls (p-value <0.05 and f-value >0.9, >2 fold change). The graph shows all significant differential abundance viruses (irrespective of fold change), with size of the viruses representing fold change (smallest – 1.00 FC, largest 2.62 FC). While there were five viruses with p-value <0.05 and f-value >0.9 in controls, average fold change was 1.09 compared to 2.23 for the nine viruses higher in ASD, suggesting ASD brains may have higher viral counts. Other – NCBI viruses, viroids, ambiguous sequences. FC – fold change.