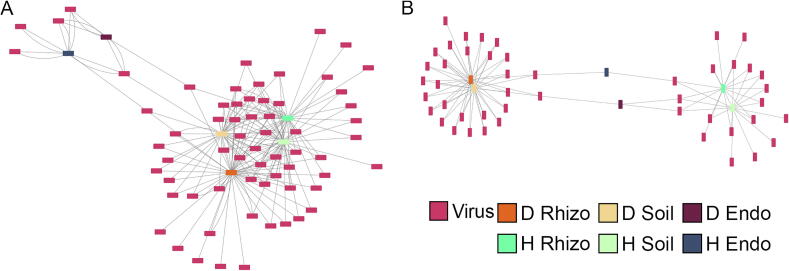
Fig. 3.
Differential abundance of viral sequences in Populus genotypes and compartments. In the A) within genotype comparison, the rhizosphere and soil samples had similar significant (p-value <0.05 and f-value >0.9) viral sequences across genotypes; however, the significant soil and rhizosphere viral sequences are more similar within genotypes than across genotypes. The two endosphere samples have few significant viral sequences, and they have more in common with each other than other compartments. In the B) between genotype comparison, the soil and rhizosphere samples for a given genotype have similar significant differentially abundant viruses. Additionally, the endosphere samples have many fewer significant differential sequences compared to the rhizosphere and soil, likely due to the overall lower abundance of viral sequences. Both graphs suggest there is a host or microbiome mediated selection of viral sequences that has some genotype and compartment specificity. D – P. deltoides, H – hybrid, Endo – endosphere, Rhizo – rhizosphere.