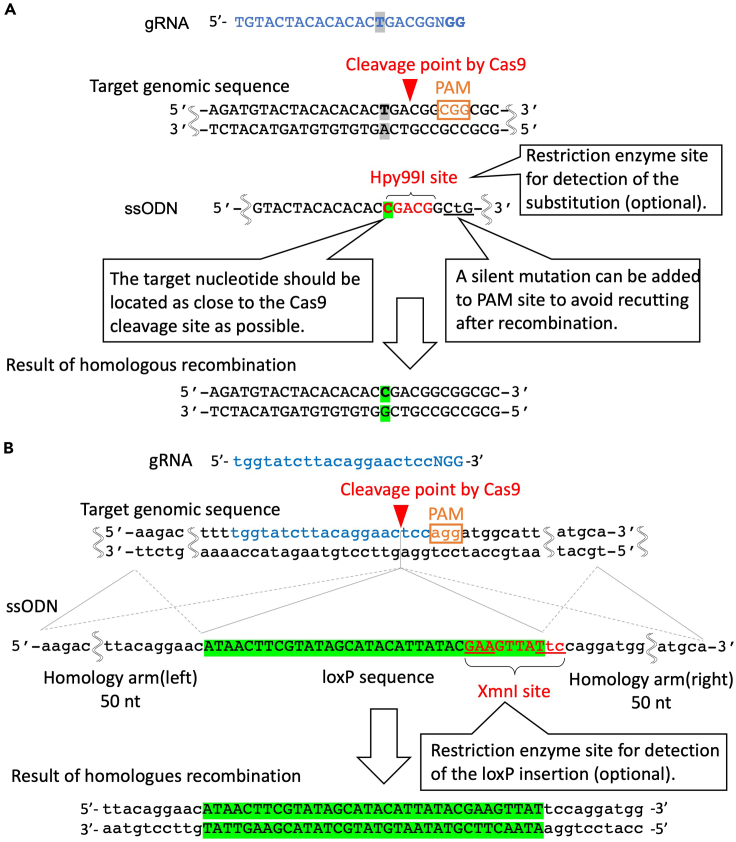
Figure 1.
gRNA and ssODN design for single nucleotide substitution and loxP sequence knockin
(A) An example of gRNA and ssODN design for single nucleotide alteration. First, the Cas9 cleavage site should be as close as the target SNP, ideally within 5 bp. ssODN template contains around 30–60 nt homology arms on both ends. In this case, the “T” base will be altered into “C” at 3-nt upstream (5′ side) from the Cas9 cleavage site. This conversion can be detected by the appearance of the Hpy99I restriction enzyme recognition site (5′-CGWCG-3′) as shown in red letters. Any other restriction enzyme site can be utilized, so long as the cleavage pattern by the restriction enzyme can be distinguished by gel electrophoresis. If it is difficult to design an appropriate restriction enzyme site after a single nucleotide alteration, the efficiency of substitution can be detected by Sanger sequencing. It is recommended to introduce a (silent) mutation at PAM or at the seed region of the target sequence to avoid recutting after recombination.
(B) gRNA targeting site should be adjusted so that the Cas9 cleavage site is located as close to the loxP insertion site as possible. When two additional nucleotide sequence (TC) is added to the 3′ end of the loxP sequence, XmnI restriction enzyme site (5′-GAANN|NNTTC-3′) can be generated, which is later utilized to assess knock-in efficiency, as described in Figure 5.
This panel is re-used from Kagita et al. (2021).