Figure 4.
Indel pattern analysis by Sanger sequencing and ICE analysis
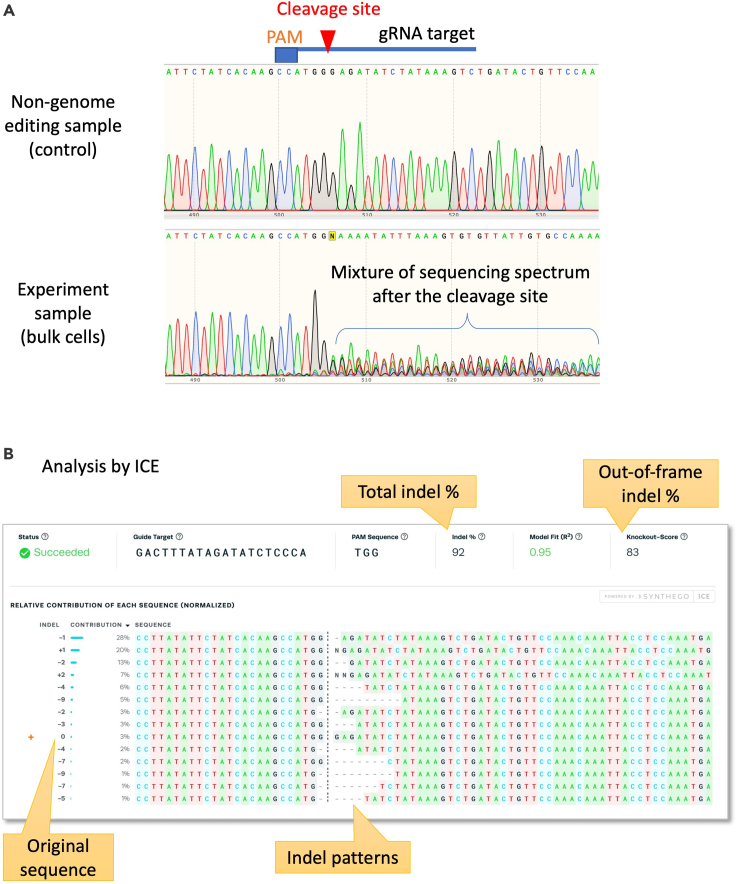
(A) Cas9 and gRNA (DMD-in55-g3-gRNA: gactttatagatatctccca tgg) complex was electroporated into Ff-XT28s05 iPSCs by using 4D-nucleofector and P4 primary cell buffer. Sequencing results of the control sample (non-transfection) and experiment sample (transfected) were shown in upper panel. In the experiment sample, a mixture of spectrum can be observed after the Cas9 cleavage site.
(B) Analysis result by ICE software. Indel percentage of the above Sanger sequencing data analyzed by ICE was 92 %, and knockout-score (exclude in-frame indels) was 83 %.