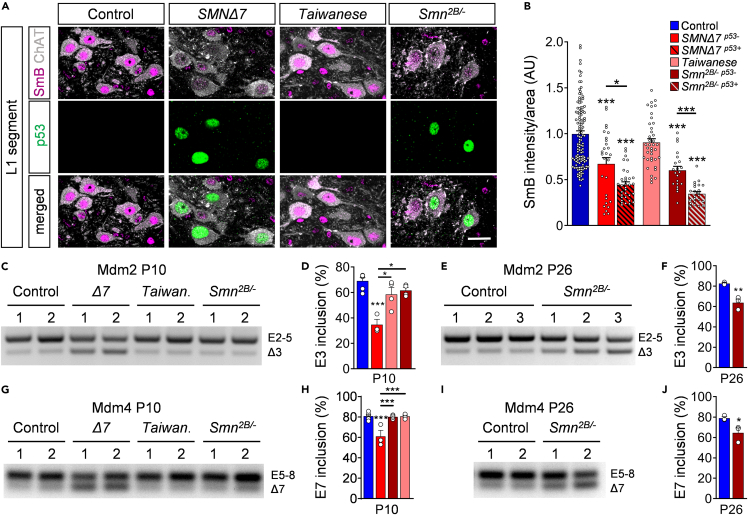
Figure 4.
Mis-splicing events of Mdm2 and Mdm4 in SMNΔ7 and Smn2B/- mice
(A) Immunostaining of ChAT+ motor neurons (gray), SmB (magenta), and p53 (green) of L1 spinal segments from control, SMNΔ7, Taiwanese, and Smn2B/- mice at P11. Scale bar = 20μm.
(B) Normalized SmB fluorescence intensity of controls, SMNΔ7 p53+, SMNΔ7 p53-, Taiwanese, and Smn2B/- p53+ and Smn2B/- p53- L1 motor neurons at P11. Each point represents SmB fluorescent intensity of a single motor neuron. Data were collected from at least three mice per genotype with pooled controls. Statistics: one-way ANOVA with Tukey’s correction. n values for motor neurons with SmB: control = 131; SMNΔ7 p53- = 28; SMNΔ7 p53+ = 33, Taiwanese = 39; Smn2B/- p53- = 23; Smn2B/- p53+ = 22.
(C) RT-PCR of Mdm2 exon 3 splicing in P10 spinal cord from the same groups as in (A).
(D) Percentage of Mdm2 exon 3 inclusions of the same groups as in (A). Statistics: one-way ANOVA with Tukey’s correction.
(E) RT-PCR of Mdm2 exon 3 splicing in P26 spinal cord from controls and Smn2B/- mice.
(F) Percentage of Mdm2 exon 3 inclusions of the same groups as in (E). Statistics: two-tailed t-test.
(G) RT-PCR of Mdm4 exon 7 splicing of spinal cords at P10 from the same groups as in (A).
(H) Percentage of Mdm4 exon 7 inclusions of P10 spinal cords from the same groups as in (A). Statistics: one-way ANOVA with Tukey’s correction.
(I) RT-PCR analysis of Mdm4 exon 7 splicing of spinal cords from P26 control and mutant Smn2B/- mice.
(J) Percentage of Mdm4 exon 7 inclusions of P26 spinal cords from the same groups as in (I). Statistics: two-tailed t-test. n values for mRNA levels for E3 and E7 inclusions: P10 control = 6; SMNΔ7 = 3; Taiwanese = 4; Smn2B/- = 4. n values for mRNA levels P26 Smn2B/- for all n = 3.
Data are presented as mean ± SEM. Asterisks on top of bars without horizontal line indicate significance compared with the control group.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.