Abstract
In recent years, regenerative medicine research using human somatic and induced pluripotent stem cells has advanced considerably, promoting clinical applications. However, it is essential that these cells are cryopreserved safely and effectively. Most cryopreservation solution agents contain dimethyl sulfoxide (DMSO), which exhibits strong toxicity and can potentially promote cell differentiation. Hence, it is important to explore substitutes for DMSO in cryoprotectant solutions. One such alternative is StemCell Keep (SCK), a DMSO-free solution that has been reported to effectively cryopreserve human induced pluripotent stem cells (hiPS cells). To clarify the effect of cryopreservation agents on cells, DNA microarray analysis is useful, as it can identify a large number of gene expression differences in cryopreserved cells, as well as functional increases in gene groups. In this study, we performed gene expression analysis of SCK-cryopreserved hiPS cells using a DNA microarray gene chip. The hiPS cells vitrified with SCK or DMSO-based vitrification solutions were thawed and cultured on Matrigel under feeder-free conditions, and RNA was extracted for DNA microarray analysis. Genes obtained from DNA microarray data were classified by the keywords of Gene Ontology Biological Process Term, and their relationships were analyzed using DAVID or the GeneMANIA database.
SCK-cryopreserved hiPS cells expressed several anti-apoptotic genes, as well as genes related to cell adhesion or proliferation at levels that were nearly equivalent to those of non-frozen hiPS cells. Gene enrichment analysis with selected genes of SCK-cryopreserved hiPS cells whose expression differences were superior to those of DAP-cryopreserved showed strong interactions of negative regulation of apoptotic process, cell adhesion and positive regulation of cell proliferation in DAVID analysis. We demonstrated that SCK successfully maintained the key functions of hiPS cells, including anti-apoptosis, cell adhesion, and cell proliferation, during cryopreservation.
Keywords: Human induced pluripotent stem cell, Cryopreservation, Vitrification, DNA microarray, Gene enrichment analysis
Highlights
-
•
Gene expression analysis of a DMSO-free solution SCK-cryopreserved hiPS was performed.
-
•
SCK-cryopreserved hiPS cells maintained anti-apoptotic, adhesion, proliferation genes.
-
•
SCK maintained the key functions of hiPS cells compared with current freezing solution.
Abbreviations
- SCK
StemCell Keep
- hiPS cells
human induced pluripotent stem cells
- hES cells
human embryonic stem cells
- CPLL
carboxylated ε-poly –l-lysine
- hPSC medium
human pluripotent stem cell medium
- Oct3/4
Octamer-binding transcription factor 3/4
- GO-BP
Biological Process Term in Gene Ontology
- DAVID
the Database for Annotation, Visualization and Integrated Discovery
- KEGG pathway
Kyoto Encyclopedia of Genes and Genomes pathway
- AP
alkaline phosphatase
- DAPI
4,6-diamidino-2-phenylindole
1. Introduction
Human embryonic stem (hES) cells and human induced pluripotent stem (hiPS) cells are used clinically and in basic medical research, as they can be differentiated into somatic cells. The differentiated somatic cells can not only be transplanted into patients, but can also be used as clinical samples for assessing various treatment options. Thus, hES and hiPS cells allow the validation of therapeutic strategies without testing them on patients directly, and promote further research on the treatment of various diseases [[1], [2], [3]]. Given their importance, large-scale cryopreservation of hES and hiPS cells for prolonged periods is essential. Additionally, it is necessary to prevent any environmental damage in the cryopreserved hES or hiPS cells because of maintaining their high multiplicity and pluripotency [4]. Hence, the development of cryopreservation agents for stem cells is essential for their storage [5,6]. Vitrification is one of the low temperature preservation methods under development for the cryopreservation of large size cells, such as oocytes and embryos [7]. This method employs rapid freezing and has been utilized for large-scale cell-construct preservation in the field of tissue engineering [[8], [9], [10]].
We previously developed a potent cryoprotective solution, StemCell Keep (SCK), for the vitrification of stem cells, and demonstrated its effectiveness using hES and hiPS cells [11,12]. SCK is composed of carboxylated ε-poly-l-lysine (CPLL), a well-known cryoprotectant; ethylene glycol; and sucrose. We found that CPLL exhibited higher cryopreservation efficiency and lower cytotoxicity than dimethyl sulfoxide (DMSO), the industry standard for cryopreservation [13,14]. Although its mechanism of protection during freezing is not explicitly clear, CPLL may protect the cell membrane at low temperatures through the suppression of ice recrystallization [15,16], as well as by providing dehydration control to inhibit intracellular ice formation [17].
In previous studies, we observed that some stem cell-maker genes were highly expressed in hiPS cells cryopreserved in SCK compared with those exposed to a DMSO-based vitrification solution [12]; furthermore, DMSO was found to enhance unexpected differentiation in stem cells. We therefore examined a wide array of gene expression in hiPS cells cryopreserved with SCK [12,18,19]. The DMSO-based vitrification solution, DAP213 solution (DAP), composed of DMSO, acetamide, and propylene glycol, was first developed for the vitrification of mouse morulae and blastocysts [20]. DAP solution has also been used for the vitrification of hES cells and hiPS cells [21].
In this study, we investigated the gene expression profiles of SCK-cryopreserved hiPS cells by using a DNA microarray gene chip and compared them with the profiles of cells preserved with DAP.
2. Materials and methods
2.1. hiPS cell culture
The hiPS cells (253G1 strain, cell passage 9–12, RIKEN BioResourse Center (Tsukuba, Japan) were maintained on a feeder layer of mitomycin C-inactivated SNL 76/7 cells (SNL cells; DS Pharma Biomedical, Osaka, Japan) [[22], [23], [24]]. SNL cells were inoculated at a density of 1.6 × 104 cells/cm2 on a 0.1% gelatin-coated 10 cm-plate and were cultured until 90% confluency. They were then mitotically inactivated by incubation with mitomycin C (10 μg/mL; Kyowa Hakko Kirin Co. Ltd., Tokyo, Japan) for 2–4 h, and were maintained on a 0.1% gelatin (Nacalai Tesque, Kyoto, Japan)-coated dish in high-glucose Dulbecco’s modified Eagle’s medium (Nacalai Tesque) containing 7% fetal bovine serum (FBS; MP Bio-Medicals, LLC Solon, OH, USA) and 1% penicillin–streptomycin (Nacalai Tesque) at 37 °C in a 5% CO2 incubator. The hiPS cells were maintained in human pluripotent stem cell (hPSC) medium (20% knockout serum replacement; Life Technologies, Carlsbad, CA, USA) containing 2 mM l-glutamine (Nacalai Tesque), 0.1 mM minimum essential medium with nonessential amino acids (Nacalai Tesque), 0.1 mM 2-mercaptoethanol (Life Technologies), and 5 ng/mL basic fibroblast growth factor (bFGF; Wako Pure Chemical, Osaka, Japan). hiPS cells were sub-cultured every 3–5 days using CTK buffer composed of 0.25% trypsin, 1 mg/ml collagenase (Life Technologies), 20% knockout serum replacement, and 1 mM CaCl2 (Nacalai Tesque) in phosphate buffer saline (PBS; Nacalai Tesque).
For feeder-free culture, hiPS cells were maintained at a density of 2 × 104 cells/cm2 on 6-cm Matrigel-coated plates (BD Biosciences, Franklin Lakes, NJ, USA) in 5 mL of SNL-conditioned medium (supernatant of SNL cells cultured in hPSC medium at a density of 2 × 104 cells/cm2 for one day, followed by further cultivation for two days before collection) containing 5 ng/mL bFGF at 37 °C in a 5% CO2 incubator.
Cells were then subcultured without the feeder layer, rinsed once with PBS, then dissociated with TrypLE Select (Life Technologies) to produce a single-cell suspension. The cells were then collected in a conical tube for centrifugation at 190g for 5 min, suspended in SNL-conditioned medium with 5 ng/mL bFGF, and plated on Matrigel-coated plates (BD Biosciences).
2.2. Vitrification and revival of hiPS cells
The vitrification solutions, SCK and DAP213 (DAP), were purchased from BioVerde (Kyoto, Japan) and Wako Pure Chemicals, respectively. For Vitrification, hiPS cells were cultured until 80% confluence, rinsed with PBS, and dissociated by treatment with CTK buffer. The detached hiPS cell colonies were gently pipetted to disperse clumps, collected in a conical tube (Thermo Fisher Scientific, Tokyo, Japan) for centrifugation at 190g for 5 min, and quickly suspended in 0.2 mL of ice-cold vitrification solution in a cryotube (Nunc). The tube was immediately immersed in liquid nitrogen and stored until required.
For the revival of vitrified hiPS cells, the cells were thawed by adding 1 mL of hPSC medium to the cryotube, and then transferred into a conical tube with 10 mL of hiPS cell medium for centrifugation at 190g for 5 min. The cells were suspended in 5 mL of SNL-conditioned medium with 5 ng/mL bFGF and 10 μM Y-27632 (Wako Pure Chemicals) onto Matrigel-coated 6-cm dishes and incubated for 24 h. After their revival, the SCK- and DAP-cryopreserved hiPS cells were cultured for one week. The medium was then replaced with SNL-conditioned medium containing bFGF only.
2.3. Alkaline phosphatase (AP) staining
A Leukocyte Alkaline Phosphatase Kit (Sigma-Aldrich, St. Louis, MO, USA) was used to assess the AP activity. Cultured hiPS cells were washed with PBS, fixed with 4% paraformaldehyde (Nacalai Tesque) for 10 min, then rinsed with distilled water. AP staining was carried according to the manufacturer’s instructions. After staining, the samples were washed with distilled water and air-dried.
2.4. Immunocytochemical analysis
The hiPS cells were cultured at the density of 1 × 105 cells/cm2 in a 24-well plate overnight and were then rinsed with PBS and fixed as described in “Alkaline phosphatase(AP) staining”. The cells were permeabilized with Perm/Wash buffer I (BD Phosflow; BD Biosciences) for 15 min. After three washes with 2% FBS in PBS, the cells were incubated with diluted primary antibodies overnight at 4 °C. The primary antibodies used were Octamer-binding transcription factor 3/4 (Oct3/4; 1:200; Santa Cruz Biotechnology, Dallas, TX, USA) and Nanog (1:200; ReproCELL, Yokohama, Japan). The cells were washed, and secondary antibodies (Goat polyclonal Ab to rabbit IgG-FITC(1:300, Santa Cruz, CA, USA) and Goat F(ab) anti-mouse IgG-FITC (1:300, Santa Cruz, CA USA)) were added to the cells for 1 h in the dark. The samples were then washed three times with 2% FBS in PBS, and one drop of mounting medium containing 4,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA, USA) was added. The cells were observed under a fluorescence microscope (Keyence, Osaka, Japan). Negative controls were prepared using the same procedure, without primary antibody treatment.
2.5. DNA microarray experiments
The SCK- or DAP-cryopreserved hiPS cells were thawed and cultured on Matrigel under feeder-free conditions until 80% confluence. The cells, at an approximate concentration of 1 × 104 cells/cm2 in a 6-cm dish, were rinsed once with PBS and dissociated using TrypLE Select (Life Technologies). Total RNA was extracted from hiPS cells using the RNeasy kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s instructions. RNA samples from one 6-cm dish per sample were then handled by the Microarray Analysis Team at Kurabo Co. Biomedical Department (Osaka, Japan) for the DNA microarray experiments. The RNA concentration and purity were first assessed. The cDNA was synthesized from the RNA samples using the Whole Transcript Sense Target Labeling Assay Schematic (Affymetrix kit; Thermo Fisher Scientific) according to manufacturer instructions. The DNA microarray gene chip, GeneChip® 3' Expression Array Service(Affymetrix), was applied using the one-color method [25], and the signal data of each probe was calculated after normalization by the robust multi-array analysis (RMA) algorithm (Affymetrix Expression Console Software v.1.0 –User Guide 2013 130–132); the sample data were directly compared. The microarray results were provided on a DNA Microarray Viewer v.1.0 (Kurabo Co. Biomedical Department). The signal log ratio data showed the difference in expression between genes of interest.
The probes, gene names, chromosomal locations, NCBI Unigene IDs, and various database IDs have been provided at:
http://www.affymetrix.com/support/technical/manual/taf_manual.affx.
The genes that exhibited larger differences in gene expression were further analyzed with respect to their gene functions and connections through Gene Ontology and KEGG in DAVID, and with respect to higher gene networks in GeneMANIA.
2.6. Enrichment analysis of genes
Genes obtained from the DNA microarray data were classified based on the keywords of Biological Process Term in Gene Ontology (GO-BP) using the DNA Microarray Viewer software. We extracted genes with the GO-BP keywords, ‘apoptosis,’ ‘cell proliferation,’ ‘cell adhesion,’ and ‘stem cell,’ and whose expression difference between SCK-cryopreserved hiPS cells and non-frozen hiPS cells or between SCK-cryopreserved hiPS cells and DAP-cryopreserved hiPS cells was 1.4-fold or 1.5-fold [26]. Selected genes were explored with respect to the biological process, molecular function, cellular component annotations, and functional relationships or clustering using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) [27,28]. The gene list of interest was uploaded to ‘DAVID Functional Annotation Bioinformatics Microarray Analysis’, and were analyzed by the ‘Functional Annotation Tool’, mainly using the categories from Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway [29,30]. The ‘Probe set ID’ of selected genes obtained from the DNA microarray data were first translated into Entrez Gene IDs using the gene conversion tool, and then introduced into the gene functional annotation tool. Finally, the gene category annotated based on the DAVID score enrichment p-value (p < 0.05) was considered.
GeneMANIA was used to identify genes related to sets of selected genes underlying specific functional themes, as identified by DNA microarray data, and the Gene Symbol of each was uploaded for analysis. The GeneMANIA algorithm comprised a linear-regression-based algorithm for calculating single, composite, functional association networks from multiple networks derived from different proteomic or genomic data sources, and for the prediction of gene function [31,32].
2.7. Statistical analysis
The counts of AP + colonies have been presented as mean ± standard deviation. Statistical analyses were performed using Excel Statistics (SSRI Co. Ltd., Tokyo, Japan). The student’s t-test was used for analyzing data when two groups were compared. Statistical significance was set at p < 0.05. In order to compare the fluctuations in gene expression in the DNA microarray data, the p-value of the t-test, Benjamini-Hochberg method, and false discovery ratio in DAVID analysis were considered.
3. Results
3.1. Vitrification of hiPS cells
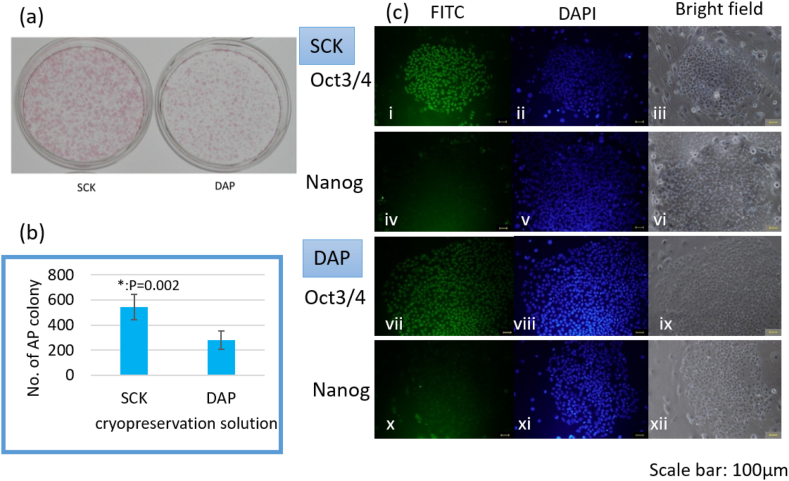
Proliferation and multipotency of SCK-cryopreserved hiPS cells for 1 week was first assessed. The number of AP + colonies generated by the SCK-cryopreserved hiPS cells (546 ± 101) was significantly higher than that of DAP-cryopreserved hiPS cells (282 ± 74; p = 0.002, Fig. 1a and b). Furthermore, the pluripotent markers Oct3/4 and Nanog [33], were found to be expressed in both SCK- and DAP-cryopreserved hiPS cells by immunocytochemical staining (Fig. 1c). Thus, the SCK-cryopreserved hiPS cells maintained their pluripotency and multipotency even after revival.
Fig. 1.
Characterization of StemCell Keep (SCK)-cryopreserved human induced pluripotent stem (hiPS) cells.
(a) Representative images of alkaline phosphatase (AP) + staining of SCK- or DAP213 (DAP)-cryopreserved hiPS cells. (b) Number of AP + colonies that generated from SCK- or DAP-cryopreserved hiPS cells. *p = 0.002 (c) Representative images of immunofluorescence staining of SCK- or DAP-cryopreserved hiPS cells. Oct 3/4-FITC staining (green), i and vii; Nanog-FITC staining (green), iv and x; DAPI, 4,6- diamidino-2-phenylindole, staining (blue), ii, v, viii and xi; bright-field images, iii, vi, ix and xii. Scale bar: 100 μm. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Enrichment analysis of ‘apoptosis’ in SCK-cryopreserved hiPS cells
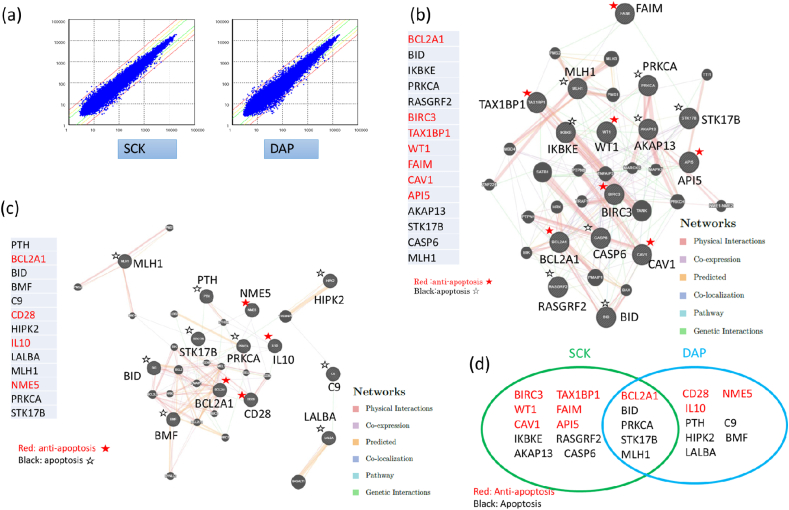
Next, we investigated differences between gene expression profiles of SCK- and DAP-cryopreserved hiPS cells and non-frozen hiPS cells, which had been maintained in normal culture conditions for at least three or four passages after reviving. The dot plots of DNA microarray data of SCK and DAP are shown in Fig. 2 and Fig. S1. The GeneChip Gene 2.0ST array probe set (Probe number: 56317) was used. We analyzed the data using DNA Microarray Viewer software, and selected four keywords—apoptosis, cell adhesion, cell proliferation, and stem cell—categorized in GO-BP, all of which were important factors for hiPS cell culture and may help further their use in research.
Fig. 2.
Annotation analysis profiles of SCK- and DAP- cryopreserved hiPS cells (I).
(a) Dot plots of SCK- and DAP- cryopreserved hiPS cells in DNA microarray data. In dot plots, red or green line upper diagonal shows 2 or 1 of signal log ratio, respectively, and those of lower diagonal shows -2 or -1 of signal log ratio, respectively. (b) GeneMANIA profile of genes of SCK-cryopreserved hiPS cells classified under ‘apoptosis’ with a difference in expression of more than 1.5-fold compared with non-frozen hiPS cells. Red font indicates anti-apoptotic genes, and black indicates apoptotic genes. (c) GeneMANIA analysis of genes in DAP-cryopreserved hiPS cells classified under ‘apoptosis’ with a difference in expression of more than 1.5-fold compared with the non-frozen hiPS cells. Red font indicates anti-apoptotic genes, and black indicates apoptotic genes. (d) Correlation diagram of genes classified under ‘apoptosis’ in Biological Process Term of Gene Ontology with a difference in expression of more than 1.5-fold between SCK- or DAP- cryopreserved cells and non-frozen hiPS cells. Red font indicates anti-apoptotic genes, and black indicates apoptotic genes. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Notably, many hiPS cells do not survive post-revival due to apoptosis [34]. Hence, we extracted apoptotic genes and assessed the difference in the expression of these genes during the freeze-thaw period. For the remaining three keywords, cell adhesion, cell proliferation, and stem cell, we compared the cryopreservation effects of SCK and DAP on hiPS cells. The genes extracted by each keyword were subjected to an enrichment analysis using DAVID, which was converted from the Probe Set IDs of DNA microarray data into Entrez Gene IDs, and categorized into Gene Ontology or KEGG pathway. Altitude gene networks of each keyword were further extracted using GeneMANIA, and their gene networks were investigated.
Table 1 shows the annotation analysis scheme of DNA microarray data (the data obtained from the microarray experiments were selected from four groups of the GO-BP by DNA Microarray Viewer), as well as the difference in gene expression between the SCK- or DAP-cryopreserved hiPS cells and non-frozen hiPS cells, or between the SCK- cryopreserved and DAP-cryopreserved hiPS cells. We extracted 15 genes using the DNA Microarray Viewer that were differentially expressed by over 1.5-fold between the SCK-cryopreserved and non-frozen hiPS cells. Among these genes, seven were categorized as anti-apoptotic (BIRC3, TAX1BP1, BCL2A1, WT1, FAIM, CAV1 and API5), while the remaining eight were categorized as apoptotic (BID, PRKCA, IKBKE, RASGRF2, STK17B, AKAP13, CASP6, and MLH1), though PRKCA exhibited both apoptotic and anti-apoptotic functions in glial cells (Table 2a).
Table 1.
Annotation analysis scheme of DNA microarray data.
| ||
| No. | Group | |
| 1 | Apoptosis | |
| 2 | Cell adhesion | |
| 3 | Cell proliferation | |
| 4 | Stem cell | |
|
||
| No. | Classification | |
| 1 | Gene expression difference between SCK- or DAP-cryopreserved hiPS cells and non-frozen hiPS cells | |
| 2 | Gene expression difference between SCK-cryopreserved hiPS cells and DAP-cryopreserved hiPS cells | |
|
||
| No. | Software | Contents |
| 1 | DNA Microarray Viewer | Signal log ratio |
| Probe set ID | ||
| 2 | Gene Ontology | Biological Process Term |
| 3 | DAVID | Entrez Gene ID |
| Net work category | ||
| KEGG | ||
| 4 | GeneMANIA | Gene network |
Table 2a.
Genes of SCK-cryopreserved hiPS cells classified under ‘apoptosis’ with a difference in expression of more than 1.5-fold compared with the non-frozen hiPS cells.
| No. | Gene Symbol | Probe Set ID | control2_Signal | SCK_Signal | SCK_Signal Log Ratio | GO Biological Process Term | Pathway Name |
|---|---|---|---|---|---|---|---|
| 1 | BCL2A1 | 16812344 | 40.4 | 77.5 | 0.94 | apoptotic process//anti-apoptosis | Apoptosis_KEGG |
| 2 | BID | 16932008 | 309.5 | 587.0 | 0.92 | induction of apoptosis via death domain receptors//glial cell apoptotic process//positive regulation of apoptotic process//neuron apoptotic process | – |
| 3 | IKBKE | 16676592 | 92.0 | 164.2 | 0.84 | DNA damage response, signal transduction resulting in induction of apoptosis | Apoptosis_KEGG |
| 4 | PRKCA | 16837128 | 270.8 | 456.4 | 0.75 | apoptotic process//cell adhesion//induction of apoptosis by extracellular signals//negative regulation of glial cell apoptotic process | G_Protein_Signaling//Wnt_signaling//Calcium_regulation_in_cardiac_cells//Smooth_muscle_contraction |
| 5 | RASGRF2 | 16997688 | 76.4 | 128.0 | 0.74 | apoptotic process | – |
| 6 | BIRC3 | 16730522 | 36.5 | 61.0 | 0.74 | apoptotic process//anti-apoptosis | – |
| 7 | TAX1BP1 | 17044568 | 644.6 | 1059.6 | 0.72 | apoptotic process//anti-apoptosis | – |
| 8 | WT1 | 16737105 | 55.6 | 87.2 | 0.65 | induction of apoptosis//negative regulation of apoptotic process | – |
| 9 | FAIM | 16946207 | 103.5 | 160.1 | 0.63 | apoptotic process//anti-apoptosis | – |
| 10 | CAV1 | 17050578 | 229.5 | 354.3 | 0.63 | induction of apoptosis by extracellular signals//positive regulation of anti-apoptosis//positive regulation of extrinsic apoptotic signaling pathway//positive regulation of intrinsic apoptotic signaling pathway | Integrin-mediated_cell_adhesion_KEGG |
| 11 | API5 | 16737482 | 60.1 | 92.2 | 0.62 | apoptotic process//anti-apoptosis | – |
| 12 | AKAP13 | 16812871 | 88.4 | 135.5 | 0.62 | apoptotic process//induction of apoptosis by extracellular signals | – |
| 13 | STK17B | 16906733 | 92.1 | 139.3 | 0.60 | apoptotic process//induction of apoptosis | – |
| 14 | CASP6 | 16978959 | 136.6 | 204.5 | 0.58 | apoptotic process//induction of apoptosis//cellular component disassembly involved in apoptotic process | – |
| 15 | MLH1 | 16938899 | 469.8 | 700.4 | 0.58 | DNA damage response, signal transduction resulting in induction of apoptosis | Ovarian_Infertility_Genes |
When these genes were assessed by DAVID under the statistical condition of p < 0.05, we found 17 terms in the GO-BP, of which the top five were ‘intrinsic apoptotic signaling pathway in response to DNA damage,’ ‘negative regulation of apoptotic process,’ ‘apoptotic process,’ ‘protein homooligomerization,’ and ‘negative regulation of necroptotic process’ (Table 2b). We also found three terms in the KEGG pathway (Table 2b). Of the total GO-BP terms, eight were related to apoptosis, while one was related with anti-apoptosis (Table 2b). In the KEGG pathway, hsa04210, three detected genes, BIRC3(IAPXIP in KEGG), BID, and CASP6, exhibited expression differences in SCK-cryopreserved hiPS cells higher than those in non-frozen hiPS cells. Because BIRC3 suppressed CASP3, CASP7, and CAPS9, which inhibit changes of substrates, it likely caused weakened apoptosis. BIRC3 is a pro-survival gene itself, and therefore suppresses overall apoptosis (Fig. S1). Furthermore, GeneMANIA analysis of the 15 genes showed that they were strongly expressed and formed one large gene network of physical interaction and pathways (Fig. 2b, Fig. S1).
Table 2b.
DAVID analysis for genes of SCK-cryopreserved hiPS cells classified under ‘apoptosis’ with a difference in expression of more than 1.5-fold compared with non-frozen hiPS cells (p < 0.05).
| No. | Category | Term | Count | % | PValue | Genes | List Total | Pop Hits | Pop Total | Fold Enrichment | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GOTERM_BP_DIRECT | GO:0008630∼intrinsic apoptotic signaling pathway in response to DNA damage | 3 | 23.1 | 3.40E-04 | IKBKE BCL2A1 MLH1 | 11 | 47 | 16792 | 97.44 | 0.079 | 0.083 | 0.082 |
| 2 | GOTERM_BP_DIRECT | GO:0043066∼negative regulation of apoptotic process | 4 | 30.8 | 0.002 | BIRC3 FAIM BCL2A1 WT1 | 11 | 455 | 16792 | 13.42 | 0.394 | 0.231 | 0.229 |
| 3 | GOTERM_BP_DIRECT | GO:0006915∼apoptotic process | 4 | 30.8 | 0.004 | BIRC3 FAIM CASP6 STK17B | 11 | 567 | 16792 | 10.77 | 0.608 | 0.231 | 0.229 |
| 4 | GOTERM_BP_DIRECT | GO:0051260∼protein homooligomerization | 3 | 23.1 | 0.005 | IKBKE CAV1 BID | 11 | 177 | 16792 | 25.87 | 0.682 | 0.231 | 0.229 |
| 5 | GOTERM_BP_DIRECT | GO:0060546∼negative regulation of necroptotic process | 2 | 15.4 | 0.005 | BIRC3 CAV1 | 11 | 8 | 16792 | 381.64 | 0.686 | 0.231 | 0.229 |
| 6 | GOTERM_BP_DIRECT | GO:0042981∼regulation of apoptotic process | 3 | 23.1 | 0.007 | BIRC3 BID CASP6 | 11 | 213 | 16792 | 21.50 | 0.807 | 0.273 | 0.271 |
| 7 | GOTERM_BP_DIRECT | GO:0043065∼positive regulation of apoptotic process | 3 | 23.1 | 0.013 | BCL2A1 BID WT1 | 11 | 300 | 16792 | 15.27 | 0.959 | 0.402 | 0.399 |
| 8 | GOTERM_BP_DIRECT | GO:0001836∼release of cytochrome c from mitochondria | 2 | 15.4 | 0.014 | BCL2A1 BID | 11 | 23 | 16792 | 132.74 | 0.964 | 0.402 | 0.399 |
| 9 | GOTERM_BP_DIRECT | GO:2001238∼positive regulation of extrinsic apoptotic signaling pathway | 2 | 15.4 | 0.015 | CAV1 BID | 11 | 26 | 16792 | 117.43 | 0.977 | 0.402 | 0.399 |
| 10 | GOTERM_BP_DIRECT | GO:0035666∼TRIF-dependent toll-like receptor signaling pathway | 2 | 15.4 | 0.017 | IKBKE BIRC3 | 11 | 28 | 16792 | 109.04 | 0.983 | 0.402 | 0.399 |
| 11 | GOTERM_BP_DIRECT | GO:2001244∼positive regulation of intrinsic apoptotic signaling pathway | 2 | 15.4 | 0.019 | CAV1 BID | 11 | 33 | 16792 | 92.52 | 0.992 | 0.430 | 0.427 |
| 12 | GOTERM_BP_DIRECT | GO:0006468∼protein phosphorylation | 3 | 23.1 | 0.029 | IKBKE PRKCA STK17B | 11 | 456 | 16792 | 10.04 | 0.999 | 0.580 | 0.576 |
| 13 | GOTERM_BP_DIRECT | GO:0001570∼vasculogenesis | 2 | 15.4 | 0.033 | CAV1 WT1 | 11 | 56 | 16792 | 54.52 | 1.000 | 0.593 | 0.588 |
| 14 | GOTERM_BP_DIRECT | GO:0031398∼positive regulation of protein ubiquitination | 2 | 15.4 | 0.037 | BIRC3 CAV1 | 11 | 64 | 16792 | 47.70 | 1.000 | 0.593 | 0.588 |
| 15 | GOTERM_BP_DIRECT | GO:0038061∼NIK/NF-kappaB signaling | 2 | 15.4 | 0.039 | IKBKE BIRC3 | 11 | 66 | 16792 | 46.26 | 1.000 | 0.593 | 0.588 |
| 16 | GOTERM_BP_DIRECT | GO:0030855∼epithelial cell differentiation | 2 | 15.4 | 0.041 | WT1 CASP6 | 11 | 70 | 16792 | 43.62 | 1.000 | 0.593 | 0.588 |
| 17 | GOTERM_BP_DIRECT | GO:0097190∼apoptotic signaling pathway | 2 | 15.4 | 0.041 | PRKCA CAV1 | 11 | 71 | 16792 | 43.00 | 1.000 | 0.593 | 0.588 |
| 18 | KEGG_PATHWAY | hsa04210:Apoptosis | 3 | 23.08 | 0.002 | BIRC3 BID CASP6 | 9 | 62 | 6879 | 36.98 | 0.163 | 0.177 | 0.177 |
| 19 | KEGG_PATHWAY | hsa05200:Pathways in cancer | 4 | 30.77 | 0.008 | BIRC3 PRKCA BID MLH1 | 9 | 393 | 6879 | 7.78 | 0.497 | 0.342 | 0.342 |
| 20 | KEGG_PATHWAY | hsa04510:Focal adhesion | 3 | 23.08 | 0.022 | BIRC3 PRKCA CAV1 | 9 | 206 | 6879 | 11.13 | 0.841 | 0.606 | 0.606 |
Next, we extracted 13 genes that demonstrated a difference in expression of more than 1.5-fold between DAP-cryopreserved and non-frozen hiPS cells. Four of these genes, BCL2A1, CD28, NME5, and IL10, were related to anti-apoptosis, while the remaining nine, BID, PRKCA, STK17B, MLH1, PTH, C9, HIPK2, BMF, and LALBA, were apoptosis-related (Table 3a). When we analyzed these genes by DAVID under the statistical condition of p < 0.05, we found ten terms in GO-BP, of which the top three were ‘GO:0032464∼positive regulation of protein homooligomerization,’ ‘GO:0043065∼positive regulation of apoptotic process,’ and ‘GO:0001836∼release of cytochrome c from mitochondria’ (Table3a, Table 3ba, b). Additionally, four terms in the KEGG pathway that were unrelated to apoptosis were noted (Table3a, Table 3ba, b). The GeneMANIA analysis of these genes revealed a network composed of ten genes showing physical interaction and pathways. However, the expression of these genes was weak, and the network was not as strong as in the SCK-cryopreserved hiPS cells (Fig. 2c). The common apoptotic genes observed in the SCK- and DAP-cryopreserved hiPS cells with a difference in expression greater than 1.5-fold were BCL2A1, BID, PRKCA, STK17B, and MLH, while the other ten genes for SCK- and eight genes for DAP-cryopreserved hiPS cells were in independent groups (Fig. 2d).
Table3a.
Genes of DAP-cryopreserved hiPS cells classified under ‘apoptosis’ with a difference in expression of more than 1.5-fold compared with non-frozen hiPS cells.
| No | Gene Symbol | Probe Set ID | control2_Signal | DAP_Signal | DAP_Signal Log Ratio | GO Biological Process Term | Pathway Name |
|---|---|---|---|---|---|---|---|
| 1 | PTH | 16735970 | 5.3 | 14.4 | 1.43 | induction of apoptosis by hormones | – |
| 2 | C9 | 16995629 | 17.7 | 36.9 | 1.06 | induction of apoptosis | Complement_Activation_Classical |
| 3 | BCL2A1 | 16812344 | 40.4 | 80.0 | 0.99 | apoptotic process//anti-apoptosis | Apoptosis_KEGG |
| 4 | PRKCA | 16837128 | 270.8 | 451.2 | 0.74 | induction of apoptosis by extracellular signals | G_Protein_Signaling//Wnt_signaling//Calcium_regulation_in_cardiac_cells//Smooth_muscle_contraction |
| 5 | CD28 | 16889807 | 24.4 | 40.4 | 0.72 | induction of apoptosis by extracellular signals//positive regulation of anti-apoptosis | Inflammatory_Response_Pathway |
| 6 | STK17B | 16906733 | 92.1 | 147.7 | 0.68 | apoptotic process//induction of apoptosis | – |
| 7 | HIPK2 | 17063461 | 676.2 | 1073.1 | 0.67 | apoptotic process//induction of apoptosis by intracellular signals//DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis//negative regulation of neuron apoptotic process | – |
| 8 | MLH1 | 16938899 | 469.8 | 741.7 | 0.66 | DNA damage response, signal transduction resulting in induction of apoptosis | Ovarian_Infertility_Genes |
| 9 | BMF | 16807324 | 95.1 | 148.4 | 0.64 | apoptotic process//induction of apoptosis by intracellular signals//activation of pro-apoptotic gene products | – |
| 10 | LALBA | 16763931 | 26.9 | 41.6 | 0.63 | induction of apoptosis | – |
| 11 | BID | 16932008 | 309.5 | 469.2 | 0.60 | apoptotic process//induction of apoptosis by intracellular signals//activation of pro-apoptotic gene products//apoptotic mitochondrial changes//glial cell apoptotic process//regulation of cell proliferation//positive regulation of apoptotic process//neuron apoptotic process//positive regulation of extrinsic apoptotic signaling pathway | – |
| 12 | NME5 | 17000342 | 22.9 | 34.4 | 0.59 | anti-apoptosis | – |
| 13 | IL10 | 16698684 | 22.2 | 33.1 | 0.58 | anti-apoptosis | – |
Table 3b.
DAVID analysis for genes of DAP-cryopreserved hiPS cells classified under ‘apoptosis’ with a difference in expression of more than 1.5-fold compared with the non-frozen hiPS cells (p < 0.05).
| No. | Category | Term | Count | % | PValue | Genes | List Total | Pop Hits | Pop Total | Fold Enrichment | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GOTERM_BP_DIRECT | GO:0032464∼positive regulation of protein homooligomerization | 2 | 15.38 | 0.005 | BID BMF | 11 | 9 | 16792 | 339.23 | 0.722 | 0.665 | 0.665 |
| 2 | GOTERM_BP_DIRECT | GO:0043065∼positive regulation of apoptotic process | 3 | 23.08 | 0.013 | BCL2A1 BID BMF | 11 | 300 | 16792 | 15.27 | 0.956 | 0.665 | 0.665 |
| 3 | GOTERM_BP_DIRECT | GO:0001836∼release of cytochrome c from mitochondria | 2 | 15.38 | 0.014 | BCL2A1 BID | 11 | 23 | 16792 | 132.74 | 0.962 | 0.665 | 0.665 |
| 4 | GOTERM_BP_DIRECT | GO:0090200∼positive regulation of release of cytochrome c from mitochondria | 2 | 15.38 | 0.017 | BID BMF | 11 | 28 | 16792 | 109.04 | 0.981 | 0.665 | 0.665 |
| 5 | GOTERM_BP_DIRECT | GO:0045944∼positive regulation of transcription from RNA polymerase II promoter | 4 | 30.77 | 0.018 | CD28 IL10 PTH HIPK2 | 11 | 981 | 16792 | 6.22 | 0.985 | 0.665 | 0.665 |
| 6 | GOTERM_BP_DIRECT | GO:1900740∼positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 2 | 15.38 | 0.018 | BID BMF | 11 | 30 | 16792 | 101.77 | 0.986 | 0.665 | 0.665 |
| 7 | GOTERM_BP_DIRECT | GO:2001244∼positive regulation of intrinsic apoptotic signaling pathway | 2 | 15.38 | 0.019 | BID BMF | 11 | 33 | 16792 | 92.52 | 0.991 | 0.665 | 0.665 |
| 8 | GOTERM_BP_DIRECT | GO:0008630∼intrinsic apoptotic signaling pathway in response to DNA damage | 2 | 15.38 | 0.028 | BCL2A1 MLH1 | 11 | 47 | 16792 | 64.96 | 0.999 | 0.761 | 0.761 |
| 9 | GOTERM_BP_DIRECT | GO:0006468∼protein phosphorylation | 3 | 23.08 | 0.029 | PRKCA HIPK2 STK17B | 11 | 456 | 16792 | 10.04 | 0.999 | 0.761 | 0.761 |
| 10 | GOTERM_BP_DIRECT | GO:0097190∼apoptotic signaling pathway | 2 | 15.38 | 0.041 | CD28 PRKCA | 11 | 71 | 16792 | 43.00 | 1.000 | 0.992 | 0.992 |
| 11 | KEGG_PATHWAY | hsa05143:African trypanosomiasis | 2 | 15.38 | 0.033 | IL10 PRKCA | 8 | 33 | 6879 | 52.11 | 0.958 | 0.887 | 0.887 |
| 12 | KEGG_PATHWAY | hsa05330:Allograft rejection | 2 | 15.38 | 0.037 | CD28 IL10 | 8 | 37 | 6879 | 46.48 | 0.971 | 0.887 | 0.887 |
| 13 | KEGG_PATHWAY | hsa04672:Intestinal immune network for IgA production | 2 | 15.38 | 0.047 | CD28 IL10 | 8 | 47 | 6879 | 36.59 | 0.989 | 0.887 | 0.887 |
| 14 | KEGG_PATHWAY | hsa05320:Autoimmune thyroid disease | 2 | 15.38 | 0.052 | CD28 IL10 | 8 | 52 | 6879 | 33.07 | 0.993 | 0.887 | 0.887 |
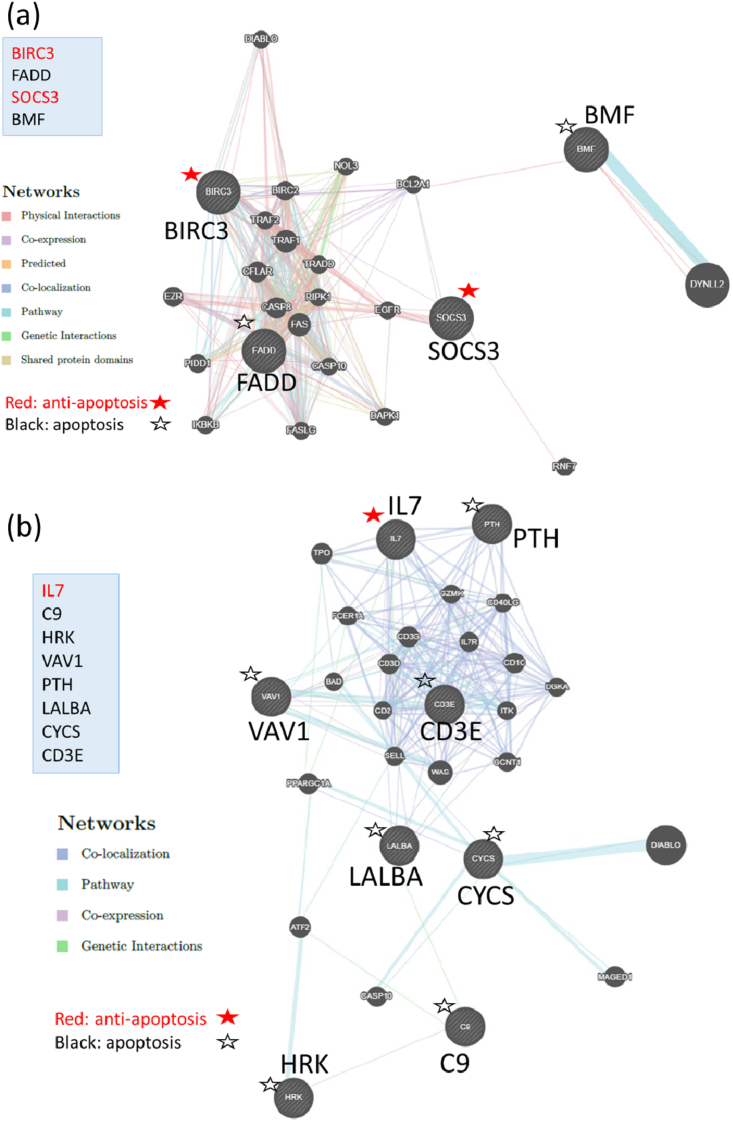
BIRC3, FADD, SOCS3, and BMF3 were expressed by SCK-cryopreserved hiPS cells and demonstrated a difference in expression of over 1.4-fold compared with the DAP-cryopreserved hiPS cells. Among these genes, BIRC3 and SOCS3 were connected to anti-apoptosis functions, while FADD and BMF3 connected to apoptosis. (Table 4a). The GeneMANIA analysis showed that BIRC3, FADD, and SOCS3 formed strong gene networks with physical interactions, co-expression, and pathways (Fig. 3a).
Table 4a.
Genes of SCK-cryopreserved hiPS cells classified under ‘apoptosis’ with a difference in expression of more than 1.4-fold when comparing with DAP-cryopreserved hiPS cells.
| No. | Gene Symbol | Probe Set ID | control2_Signal | SCK_Signal | SCK_Signal Log Ratio | DAP_Signal | DAP_Signal Log Ratio | Difference | GO Biological Process Term | Pathway Name |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | BIRC3 | 16730522 | 36.5 | 61.0 | 0.739 | 29.7 | −0.30 | 1.04 | apoptotic process//anti-apoptosis | |
| 2 | FADD | 16741426 | 219.9 | 277.6 | 0.336 | 187.9 | −0.23 | 0.56 | apoptotic process//activation of cysteine-type endopeptidase activity involved in apoptotic process//induction of apoptosis by extracellular signals//induction of apoptosis via death domain receptors//activation of pro-apoptotic gene products//positive regulation of apoptotic process//extrinsic apoptotic signaling pathway | Apoptosis//Apoptosis_GenMAPP//Apoptosis_KEGG |
| 3 | SOCS3 | 17117736 | 72.1 | 84.4 | 0.228 | 58.2 | −0.31 | 0.54 | anti-apoptosis | |
| 4 | BMF | 16799423 | 24.9 | 35.9 | 0.527 | 25.0 | 0.01 | 0.52 | apoptotic process//induction of apoptosis by intracellular signals//activation of pro-apoptotic gene products ] |
Fig. 3.
Annotation analysis profiles of SCK- and DAP- cryopreserved hiPS cells (II). (a) GeneMANIA profile of genes of SCK-cryopreserved hiPS cells classified under ‘apoptosis’ with a difference in expression of more than 1.4-fold compared with the DAP-cryopreserved hiPS cells. (b) GeneMANIA profile of genes of DAP-cryopreserved hiPS cells classified under ‘apoptosis’ with a difference in expression of more than 1.4-fold compared with the SCK-cryopreserved hiPS cells. Red font indicates anti-apoptotic genes, and black indicates apoptotic genes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In contrast, IL7, PTH, VAV1, LALBA, CD3E, HRK, CYCS, and C9 were expressed by DAP-cryopreserved hiPS cells and a difference in expression of over 1.4-fold compared with the SCK-cryopreserved hiPS cells. Among these genes, only IL7 was associated with an anti-apoptotic function. We further analyzed these genes by DAVID, and extracted five terms in GO-BP and two terms in the KEGG pathway (Table 4a, Table 4b, Table 4ca–c). The GeneMANIA analysis demonstrated that four of these genes, IL7, PTH, VAV1, and CD3E, formed a strong gene network with co-localization, co-expression, and pathways (Fig. 3b). Thus, our data indicated that the genes classified under ‘apoptosis’ in SCK-cryopreserved hiPS cells formed a strong gene network with both apoptotic and anti-apoptotic functions.
Table 4b.
Genes of DAP-cryopreserved hiPS cells classified under ‘apoptosis’ with a difference in expression of more than 1.4-fold, when comparing between SCK-cryopreserved and DAP-cryopreserved hiPS cells.
| No. | Gene Symbol | Probe Set ID | control2_Signal | SCK_Signal | SCK_Signal Log Ratio | DAP_Signal | DAP_Signal Log Ratio | Difference | GO Biological Process Term | Pathway Name |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PTH | 16735970 | 5.3 | 7.5 | 0.49 | 14.4 | 1.43 | 0.94 | induction of apoptosis by hormones | – |
| 2 | IL7 | 17078434 | 23.8 | 17.2 | −0.47 | 29.6 | 0.31 | 0.78 | anti-apoptosis | – |
| 3 | VAV1 | 16857490 | 87.9 | 61.2 | −0.52 | 98.8 | 0.17 | 0.69 | apoptotic process | – |
| 4 | LALBA | 16763931 | 26.9 | 27.2 | 0.02 | 41.6 | 0.63 | 0.61 | induction of apoptosis | – |
| 5 | CD3E | 16731795 | 36.9 | 33.6 | −0.14 | 49.3 | 0.42 | 0.55 | induction of apoptosis by extracellular signals//regulation of apoptotic process | – |
| 6 | HRK | 16770799 | 70.3 | 42.1 | −0.74 | 61.4 | −0.20 | 0.54 | apoptotic process//induction of apoptosis//positive regulation of apoptotic process//positive regulation of neuron apoptotic process | – |
| 7 | CYCS | 17055970 | 159.6 | 90.6 | −0.82 | 131.1 | −0.28 | 0.53 | apoptotic DNA fragmentation//apoptotic process//induction of apoptosis by intracellular signals//activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c | Apoptosis//Apoptosis_GenMAPP//Apoptosis_KEGG |
| 8 | C9 | 16995629 | 17.7 | 26.0 | 0.56 | 36.9 | 1.06 | 0.50 | induction of apoptosis//activation of cysteine-type endopeptidase activity involved in apoptotic process | Complement_Activation_Classical |
Table 4c.
DAVID analysis for genes of DAP-cryopreserved hiPS cells classified under “apoptosis” with a difference in expression of more than 1.4-fold, when compared with SCK-cryopreserved hiPS cells (p < 0.05).
| No | Category | Term | Count | % | PValue | Genes | List Total | Pop Hits | Pop Total | Fold Enrichment | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GOTERM_BP_DIRECT | GO:0045453∼bone resorption | 2 | 28.6 | 0.005 | PTH IL7 | 5 | 22 | 16792 | 305.3 | 0.363 | 0.296 | 0.296 |
| 2 | GOTERM_BP_DIRECT | GO:0048873∼homeostasis of number of cells within a tissue | 2 | 28.6 | 0.007 | PTH IL7 | 5 | 29 | 16792 | 231.6 | 0.448 | 0.296 | 0.296 |
| 3 | GOTERM_BP_DIRECT | GO:0007186∼G-protein coupled receptor signaling pathway | 3 | 42.9 | 0.02 | PTH CD3E VAV1 | 5 | 899 | 16792 | 11.2 | 0.75 | 0.397 | 0.397 |
| 4 | GOTERM_BP_DIRECT | GO:0031295∼T cell costimulation | 2 | 28.6 | 0.02 | CD3E VAV1 | 5 | 78 | 16792 | 86.1 | 0.798 | 0.397 | 0.397 |
| 5 | GOTERM_BP_DIRECT | GO:0010468∼regulation of gene expression | 2 | 28.6 | 0.02 | PTH IL7 | 5 | 100 | 16792 | 67.2 | 0.872 | 0.406 | 0.406 |
| 6 | KEGG_PATHWAY | hsa04640:Hematopoietic cell lineage | 2 | 28.6 | 0.03 | IL7 CD3E | 3 | 87 | 6879 | 52.7 | 0.368 | 0.26 | 0.26 |
| 7 | KEGG_PATHWAY | hsa04660:T cell receptor signaling pathway | 2 | 28.6 | 0.03 | CD3E VAV1 | 3 | 100 | 6879 | 45.9 | 0.41 | 0.26 | 0.26 |
3.3. Enrichment analysis of ‘cell adhesion,’ ‘cell proliferation,’ and ‘stem cell’ in SCK-cryopreserved hiPS cells
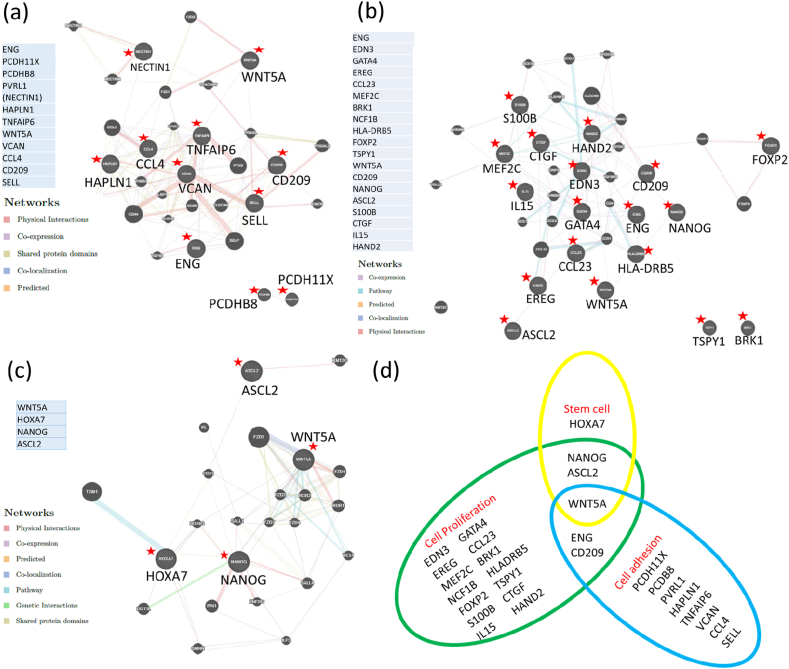
During the revival and subsequent culture period, cryopreserved hiPS cells are exposed to environmental stress, and must maintain sufficient proliferation, adhesion, and stemness. Hence, we selected three keywords, ‘cell adhesion,’ ‘cell proliferation,’ and ‘stem cell’ in GO-BP from the DNA microarray data and examined the genes of SCK-cryopreserved hiPS cells that exhibited a difference in expression of over 1.4-fold compared with the DAP-cryopreserved hiPS cells (Table 1). With respect to ‘cell adhesion,’ 11 genes (ENG, PCDH11X, PCDHB8, PVRL1, HAPLN1, TNFAIP6, WNT5A, VCAN, CCL4, CD209, and SELL) were extracted. Apart from VCAN and CD209, the differences in gene expression in the DAP-cryopreserved hiPS cells were lower than those in the non-frozen hiPS cells. Additionally, the number of SCK-cryopreserved hiPS cells that survived post revival was higher than that of DAP-cryopreserved hiPS cells. Furthermore, genes such as HAPLN1, TNFAIP6, and CCL4 of DAP-cryopreserved hiPS cells were expressed at lower levels than those of in non-frozen cells.
The average signal log ratio of 11 genes in the SCK-cryopreserved hiPS cells was 0.28 ± 0.37, and was significantly higher than that of DAP-cryopreserved hiPS cells (−0.37 ± 0.36; p = 4.1 × 10-8; Table 5a). When we analyzed these genes by DAVID, we found five terms in GO-BP, of which the top three were ‘GO:0007155∼cell adhesion,’ ‘GO:0007157∼heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules,’ and ‘GO:0071346∼cellular response to interferon-gamma’ (Table 5b). Additionally, the GeneMANIA analysis demonstrated that, although the gene expression was not strong, there were gene networks formed between HAPLN1, TNFAIP6, VCAN, CCL4, CD209, and SELL with co-expression and pathways (Fig. 4a).
Table 5a.
Genes of SCK-cryopreserved hiPS cells classified under ‘cell adhesion’ with a difference in expression of more than 1.4-fold when compared with DAP-cryopreserved hiPS cells.
| No. | Gene Symbol | Probe Set ID | control2_Signal | SCK_Signal | SCK_Signal Log Ratio | DAP_Signal | DAP_Signal Log Ratio | Difference | GO Biological Process Term |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ENG | 17098594 | 255.7 | 380.6 | 0.57 | 193.7 | −0.4 | 0.97 | cell adhesion |
| 2 | PCDH11X | 17105249 | 198.4 | 279.3 | 0.49 | 144.2 | −0.46 | 0.95 | homophilic cell adhesion |
| 3 | PCDHB8 | 16990284 | 30.3 | 33.1 | 0.13 | 21 | −0.53 | 0.66 | homophilic cell adhesion |
| 4 | PVRL1 (NECTIN1) | 16745380 | 112 | 128.5 | 0.2 | 83 | −0.43 | 0.63 | cell adhesion//homophilic cell adhesion//heterophilic cell-cell adhesion//cell-cell adhesion//adherens junction organization |
| 5 | HAPLN1 | 16997802 | 75.2 | 73.8 | −0.03 | 48.6 | −0.63 | 0.6 | cell adhesion |
| 6 | TNFAIP6 | 16886491 | 584.6 | 458 | −0.35 | 305.2 | −0.94 | 0.59 | cell adhesion |
| 7 | WNT5A | 16955197 | 118.4 | 119.6 | 0.01 | 80 | −0.57 | 0.58 | positive regulation of cell-cell adhesion mediated by cadherin |
| 8 | VCAN | 16997799 | 102.6 | 202.2 | 0.98 | 136.9 | 0.42 | 0.56 | cell adhesion |
| 9 | CCL4 | 16833420 | 41.4 | 40.8 | −0.02 | 27.8 | −0.57 | 0.55 | cell adhesion |
| 10 | CD209 | 16868000 | 95.9 | 149.2 | 0.64 | 102 | 0.09 | 0.55 | heterophilic cell-cell adhesion//leukocyte cell-cell adhesion |
| 11 | SELL | 16696237 | 34.9 | 49.5 | 0.5 | 34.1 | −0.03 | 0.54 | cell adhesion |
Table 5b.
DAVID analysis for genes of SCK-cryopreserved hiPS cells classified under ‘cell adhesion’ with a difference in expression of more than 1.4-fold when compared with DAP-cryopreserved hiPS cells (p < 0.05).
| No. | Category | Term | Count | % | PValue | Genes | List Total | Pop Hits | Pop Total | Fold Enrichment | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GOTERM_BP_DIRECT | GO:0007155∼cell adhesion | 4 | 40 | 3.82E-04 | CCL4 PVRL1 SELL TNFAIP6 | 7 | 459 | 16792 | 20.91 | 0.068 | 0.07 | 0.07 |
| 2 | GOTERM_BP_DIRECT | GO:0007157∼heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 2 | 20 | 0.02 | PVRL CD209 | 7 | 50 | 16792 | 95.95 | 0.963 | 1 | 1 |
| 3 | GOTERM_BP_DIRECT | GO:0071346∼cellular response to interferon-gamma | 2 | 20 | 0.02 | CCL4 WNT5A | 7 | 57 | 16792 | 84.17 | 0.977 | 1 | 1 |
| 4 | GOTERM_BP_DIRECT | GO:0050729∼positive regulation of inflammatory response | 2 | 20 | 0.03 | CCL4 WNT5A | 7 | 73 | 16792 | 65.72 | 0.992 | 1 | 1 |
| 5 | GOTERM_BP_DIRECT | GO:0046718∼viral entry into host cell | 2 | 20 | 0.03 | PVRL CD209 | 7 | 80 | 16792 | 59.97 | 0.995 | 1 | 1 |
| 6 | INTERPRO | IPR016186:C-type lectin-like | 3 | 30 | 4.60E-04 | CD209 SELL TNFAIP6 | 7 | 104 | 18559 | 76.48 | 0.012 | 0.007 | 0.007 |
| 7 | INTERPRO | IPR016187:C-type lectin fold | 3 | 30 | 5.33E-04 | CD209 SELL TNFAIP6 | 7 | 112 | 18559 | 71.02 | 0.014 | 0.007 | 0.007 |
| 8 | INTERPRO | IPR018378:C-type lectin, conserved site | 2 | 20 | 0.01 | CD209 SELL | 7 | 44 | 18559 | 120.51 | 0.309 | 0.123 | 0.123 |
| 9 | INTERPRO | IPR001304:C-type lectin | 2 | 20 | 0.03 | CD209 SELL | 7 | 89 | 18559 | 59.58 | 0.528 | 0.185 | 0.185 |
| 10 | SMART | SM00034:CLECT | 2 | 20 | 0.05 | CD209 SELL | 7 | 86 | 10057 | 33.41 | 0.433 | 0.553 | 0.553 |
Fig. 4.
Annotation analysis profiles of SCK- or DAP- cryopreserved hiPS cells (III). (a) GeneMANIA profile of genes of SCK-cryopreserved hiPS cells classified under ‘cell adhesion’ that exhibited a difference in expression larger than that of DAP-cryopreserved hiPS cells. (b) GeneMANIA profile of genes of SCK-cryopreserved hiPS cells classified under ‘cell proliferation’ with a difference in expression of more than 1.4-fold compared with the DAP-cryopreserved hiPS cells. (c) GeneMANIA profile of genes of SCK-cryopreserved hiPS cells classified under ‘stem cell’ with a difference in expression of more than 1.4-fold compared with the DAP-cryopreserved hiPS cells. Red stars indicate selected genes in a, b and c. (d) Gene expression profile of SCK-cryopreserved hiPS cells that exhibited a difference in expression of more than 1.4-fold compared with the DAP-cryopreserved hiPS cells. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
With respect to ‘cell proliferation,’ 19 genes (ENG, EDN3, GATA4, EREG, CCL23, MEF2C, BRK1, NCF1B, HLS-DRB5, FOXP2, TSPY1, WNT5A, CD209, NANOG, ASCL2, S100B, CTGF, IL15, and HAND2) of SCK-cryopreserved hiPS cells were found to exhibit a difference in expression of over 1.4-fold compared with the DAP-cryopreserved hiPS cells. Notably, the signal log ratios of DAP-cryopreserved hiPS cells were found to be negative in all but three genes: BRK1, CD209, and S100B. In contrast, the differences in gene expression of SCK-cryopreserved hiPS cells were mostly positive, except for three genes: FOXP2, ASCL2, and IL15. The average signal log ratio of the 19 genes of SCK-cryopreserved hiPS cells was 0.30 ± 0.31, which was significantly higher than that of DAP-cryopreserved hiPS cells (−0.36 ± 0.27; p = 1.3 × 10-14; Table 6a). When we assessed these genes by DAVID, we found 17 terms in GO-BP, of which the top three were ‘GO:0007267∼cell-cell signaling,’ ‘GO:0008284∼positive regulation of cell proliferation,’ and ‘GO:0001947∼heart looping,’ and two terms in the KEGG pathway (Table 6b). The GeneMANIA analysis showed one gene network group made up of 13 genes, apart from BRK1, FOXP2, TSPY1, and ASCL2, with co-expression and pathways. However, their pathways and physical interactions were found to be weak (Fig. 4b).
Table 6a.
Genes of SCK-cryopreserved hiPS cells classified under ‘cell proliferation’ with a difference in expression of more than 1.4-fold when compared with DAP-cryopreserved hiPS cells.
| No. | Gene Symbol | Probe Set ID | control2_Signal | SCK_Signal | SCK_Signal Log Ratio | DAP_Signal | DAP_Signal Log Ratio | difference | GO Biological Process Term |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ENG | 17098594 | 255.7 | 380.6 | 0.57 | 193.7 | −0.4 | 0.97 | regulation of cell proliferation |
| 2 | EDN3 | 16915412 | 68.6 | 115.2 | 0.75 | 65.1 | −0.08 | 0.82 | positive regulation of cell proliferation |
| 3 | GATA4 | 17065780 | 50.7 | 68.8 | 0.44 | 39.1 | −0.37 | 0.81 | positive regulation of cardiac muscle cell proliferation |
| 4 | EREG | 16967843 | 38.4 | 55.3 | 0.52 | 31.7 | −0.28 | 0.8 | positive regulation of cell proliferation//negative regulation of cell proliferation//keratinocyte proliferation//positive regulation of smooth muscle cell proliferation//negative regulation of epithelial cell proliferation//negative regulation of smooth muscle cell differentiation |
| 5 | CCL23 | 16843567 | 36.2 | 50.7 | 0.49 | 29.3 | −0.31 | 0.79 | negative regulation of cell proliferation |
| 6 | MEF2C | 16997953 | 34.7 | 41.7 | 0.27 | 25.3 | −0.46 | 0.72 | positive regulation of B cell proliferation//muscle cell differentiation//positive regulation of cardiac muscle cell proliferation//epithelial cell proliferation involved in renal tubule morphogenesis |
| 7 | BRK1 | 17118001 | 105.1 | 177.5 | 0.76 | 107.7 | 0.03 | 0.72 | positive regulation of cell proliferation |
| 8 | NCF1B | 17046911 | 24.7 | 33.3 | 0.43 | 20.3 | −0.28 | 0.71 | cell proliferation |
| 9 | HLA-DRB5 | 17017885 | 14.8 | 15.2 | 0.04 | 9.8 | −0.59 | 0.63 | negative regulation of T cell proliferation |
| 10 | FOXP2 | 17050455 | 46.8 | 41.6 | −0.17 | 27 | −0.79 | 0.62 | positive regulation of mesenchymal cell proliferation//positive regulation of epithelial cell proliferation involved in lung morphogenesis |
| 11 | TSPY1 | 17116131 | 50.4 | 60.9 | 0.27 | 39.6 | −0.35 | 0.62 | cell proliferation |
| 12 | WNT5A | 16955197 | 118.4 | 119.6 | 0.01 | 80 | −0.57 | 0.58 | positive regulation of endothelial cell proliferation//positive regulation of mesenchymal cell proliferation//epithelial cell proliferation involved in mammary gland duct elongation//hemopoietic stem cell proliferation//negative regulation of mesenchymal cell proliferation |
| 13 | CD209 | 16868000 | 95.9 | 149.2 | 0.64 | 102 | 0.09 | 0.55 | regulation of T cell proliferation |
| 14 | NANOG | 16747852 | 481.4 | 507.4 | 0.08 | 349 | −0.46 | 0.54 | cell proliferation |
| 15 | ASCL2 | 16734420 | 95.7 | 74.9 | −0.35 | 51.7 | −0.89 | 0.54 | negative regulation of Schwann cell proliferation |
| 16 | S100B | 16926754 | 52.7 | 82.1 | 0.64 | 56.7 | 0.11 | 0.54 | cell proliferation |
| 17 | CTGF | 17118180 | 476.1 | 616.9 | 0.37 | 425.8 | −0.16 | 0.53 | positive regulation of cell proliferation |
| 18 | IL15 | 16970971 | 41 | 40 | −0.04 | 27.8 | −0.56 | 0.52 | NK T cell proliferation//positive regulation of cell proliferation//positive regulation of NK cell proliferation//positive regulation of T cell proliferation//negative regulation of smooth muscle cell proliferation |
| 19 | HAND2 | 16981542 | 86.1 | 90.6 | 0.07 | 63.3 | −0.44 | 0.52 | regulation of secondary heart field cardioblast proliferation//mesenchymal cell proliferation |
Table 6b.
DAVID analysis for genes of SCK-cryopreserved hiPS cells classified under ‘cell proliferation’ with a difference in expression of more than 1.4-fold when compared with DAP-cryopreserved hiPS cells (p < 0.05).
| No. | Category | Term | Count | % | PValue | Genes | List Total | Pop Hits | Pop Total | Fold Enrichment | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GOTERM_BP_DIRECT | GO:0007267∼cell-cell signaling | 5 | 27.8 | 4.54E-05 | IL15 EREG GATA CCL23 EDN3 | 15 | 254 | 16792 | 22.04 | 0.016 | 0.017 | 0.017 |
| 2 | GOTERM_BP_DIRECT | GO:0008284∼positive regulation of cell proliferation | 5 | 27.8 | 4.70E-04 | S100B IL15 EREG EDN3 BRK1 | 15 | 466 | 16792 | 12.01 | 0.158 | 0.086 | 0.086 |
| 3 | GOTERM_BP_DIRECT | GO:0001947∼heart looping | 3 | 16.7 | 0.001 | WNT5A HAND2 GATA4 | 15 | 61 | 16792 | 55.06 | 0.343 | 0.14 | 0.14 |
| 4 | GOTERM_BP_DIRECT | GO:0050729∼positive regulation of inflammatory response | 3 | 16.7 | 0.002 | WNT5A IL15 CCL23 | 15 | 73 | 16792 | 46.01 | 0.452 | 0.15 | 0.15 |
| 5 | GOTERM_BP_DIRECT | GO:0045840∼positive regulation of mitotic nuclear division | 2 | 11.1 | 0.021 | EREG EDN3 | 15 | 26 | 16792 | 86.11 | 1 | 1 | 1 |
| 6 | GOTERM_BP_DIRECT | GO:0002053∼positive regulation of mesenchymal cell proliferation | 2 | 11.1 | 0.021 | WNT5A FOXP2 | 15 | 26 | 16792 | 86.11 | 1 | 1 | 1 |
| 7 | GOTERM_BP_DIRECT | GO:0008283∼cell proliferation | 3 | 16.7 | 0.036 | S100B TSPY1 NANOG | 15 | 366 | 16792 | 9.18 | 1 | 1 | 1 |
| 8 | GOTERM_BP_DIRECT | GO:0045165∼cell fate commitment | 2 | 11.1 | 0.038 | WNT5A GATA4 | 15 | 46 | 16792 | 48.67 | 1 | 1 | 1 |
| 9 | GOTERM_BP_DIRECT | GO:0048146∼positive regulation of fibroblast proliferation | 2 | 11.1 | 0.044 | WNT5A EREG | 15 | 54 | 16792 | 41.46 | 1 | 1 | 1 |
| 10 | GOTERM_BP_DIRECT | GO:0045944∼positive regulation of transcription from RNA polymerase II promoter | 4 | 22.2 | 0.045 | WNT5A HAND2 NANOG GATA4 | 15 | 981 | 16792 | 4.56 | 1 | 1 | 1 |
| 11 | GOTERM_BP_DIRECT | GO:0019882∼antigen processing and presentation | 2 | 11.1 | 0.045 | HLA-DRB5 CD209 | 15 | 55 | 16792 | 40.71 | 1 | 1 | 1 |
| 12 | GOTERM_BP_DIRECT | GO:0050680∼negative regulation of epithelial cell proliferation | 2 | 11.1 | 0.046 | WNT5A EREG | 15 | 56 | 16792 | 39.98 | 1 | 1 | 1 |
| 13 | GOTERM_BP_DIRECT | GO:0042733∼embryonic digit morphogenesis | 2 | 11.1 | 0.046 | WNT5A HAND2 | 15 | 56 | 16792 | 39.98 | 1 | 1 | 1 |
| 14 | GOTERM_BP_DIRECT | GO:0071346∼cellular response to interferon-gamma | 2 | 11.1 | 0.047 | WNT5A CCL23 | 15 | 57 | 16792 | 39.28 | 1 | 1 | 1 |
| 15 | GOTERM_BP_DIRECT | GO:0006955∼immune response | 3 | 16.7 | 0.047 | IL15 HLA-DRB5 CCL23 | 15 | 421 | 16792 | 7.98 | 1 | 1 | 1 |
| 16 | GOTERM_BP_DIRECT | GO:0035019∼somatic stem cell population maintenance | 2 | 11.1 | 0.05 | ASCL2 NANOG | 15 | 65 | 16792 | 34.45 | 1 | 1 | 1 |
| 17 | GOTERM_BP_DIRECT | GO:0030593∼neutrophil chemotaxis | 2 | 11.1 | 0.05 | CCL23 EDN3 | 15 | 66 | 16792 | 33.92 | 1 | 1 | 1 |
| 18 | KEGG_PATHWAY | hsa05166:HTLV-I infection | 3 | 16.7 | 0.033 | WNT5A IL15 HLA-DRB5 | 9 | 254 | 6879 | 9.03 | 0.719 | 0.999 | 0.999 |
| 19 | KEGG_PATHWAY | hsa04672:Intestinal immune network for IgA production | 2 | 11.1 | 0.053 | IL15, HLA-DRB5 | 9 | 47 | 6879 | 32.52 | 0.876 | 0.999 | 0.999 |
Finally, we extracted four genes of SCK-cryopreserved hiPS cells that presented a difference in expression of over 1.4-fold compared with the DAP-cryopreserved hiPS cells, WNT5A, HOXA7, NANOG, and ASCL2. The average signal log ratio of these genes in SCK-cryopreserved hiPS cells was -0.03 ± 0.19, which was significantly higher than that of DAP-cryopreserved hiPS cells (−0.58 ± 0.19; p = 7.4 × 10-6; Table 7a). Finally, when these genes were analyzed by DAVID, we found two terms in the GO-BP: ‘GO:0045944∼positive regulation of transcription from RNA polymerase II promoter’ and ‘GO:0035019∼somatic stem cell population maintenance’ (Table 7a, Table 7ba, b). The GeneMANIA analysis demonstrated a pattern of strong gene expression and gene networks formed by three genes with physical interaction, co-expression, and pathways (Fig. 4c).
Table 7a.
Genes of SCK-cryopreserved hiPS cells classified under ‘stem cell’ with a difference in expression of more than 1.4-fold when compared with DAP-cryopreserved hiPS cells.
| No. | Gene Symbol | Probe Set ID | control2_Signal | SCK_Signal | SCK_Signal Log Ratio | DAP_Signal | DAP_Signal Log Ratio | Difference | GO Biological Process Term |
|---|---|---|---|---|---|---|---|---|---|
| 1 | WNT5A | 16955197 | 118.4 | 119.6 | 0.01 | 80 | −0.57 | 0.58 | hemopoietic stem cell proliferation |
| 2 | HOXA7 | 17056152 | 91.2 | 101.2 | 0.15 | 69.6 | −0.39 | 0.54 | stem cell differentiation |
| 3 | NANOG | 16747852 | 481.4 | 507.4 | 0.08 | 349 | −0.46 | 0.54 | somatic stem cell maintenance |
| 4 | ASCL2 | 16734420 | 95.7 | 74.9 | −0.35 | 51.7 | −0.89 | 0.54 | somatic stem cell maintenance |
Table 7b.
DAVID analysis of genes of SCK-cryopreserved hiPS cells classified under ‘stem cell’ with a difference in expression of more than 1.4-fold when compared with DAP-cryopreserved hiPS cells.
| No | Category | Term | Count | % | PValue | Genes | List Total | Pop Hits | Pop Total | Fold Enrichment | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GOTERM_BP_DIRECT | GO:0045944∼positive regulation of transcription from RNA polymerase II promoter | 3 | 75 | 0.01 | WNT5A HOXA7 NANOG | 4 | 981 | 16792 | 12.84 | 0.784 | 0.897 | 0.897 |
| 2 | GOTERM_BP_DIRECT | GO:0035019∼somatic stem cell population maintenance | 2 | 50 | 0.01 | ASCL2 NANOG | 4 | 65 | 16792 | 129.17 | 0.835 | 0.897 | 0.897 |
| 3 | KEGG_PATHWAY | hsa04550:Signaling pathways regulating pluripotency of stem cells | 2 | 50 | 0.02 | WNT5A NANOG | 2 | 140 | 6879 | 49.14 | 0.152 | 0.116 | 0.116 |
| 4 | KEGG_PATHWAY | hsa05205:Proteoglycans in cancer | 2 | 50 | 0.03 | WNT5A NANOG | 2 | 200 | 6879 | 34.39 | 0.21 | 0.116 | 0.116 |
The inclusion diagram of genes of SCK-cryopreserved hiPS cells with higher expression than those of DAP-cryopreserved hiPS cells showed that NANOG, ASCL2, ENG, and CD209 were stronger in two functions, while WNT5A appeared in three (Fig. 4d). Additionally, the genes BIRC3, BID and CASP6, classified under the keyword ‘apoptosis’ in SCK-cryopreserved hiPS cells and with a difference in expression 1.5-fold greater than that in non-frozen hiPS cells, were also categorized under ‘focal adhesion’ in the KEGG pathway, and were associated with cell-cell adhesion and followed cell survival (Table 2(a) No.1,4,10 and Table 2(b) No.20; Fig. S2).
These findings suggest that SCK-cryopreserved hiPS cells would be more quickly cultured and maintained in good condition after thawing compared to DAP-cryopreserved cells.
4. Discussion
There are two cryopreservation methods for stem cells: vitrification and slow freezing [35,36]. Although the freezing volume used for the vitrification of cells is smaller, the damage incurred during freeze-thaw is less than that incurred during slow freezing [[37], [38], [39], [40]]. We previously demonstrated that SCK, a DMSO-free cryopreservation solution, exhibited excellent cryoprotectant properties, especially for the preservation of hiPS cells and hES cells by vitrification. In this study, we reported that SCK-cryopreserved hiPS cells retained their multipotency and pluripotency post-revival [11,12].
After revival, the SCK-cryopreserved hiPS cells were found to proliferate faster and with higher potency when compared with the DAP-cryopreserved hiPS cells (Fig. 1). Notably, when DAP-cryopreserved hiPS cells were thawed and cultured, a large number of cells did not survive the freeze-thaw process. The few surviving cells required a long period of culture in order to generate cells in sufficient numbers to perform an experiment. In contrast, the SCK-cryopreserved hiPS cells exhibited strong adhesive properties to the culture dish and proliferated quickly, even though some cells did not survive the freeze-thaw process. Thus, we concluded that SCK-treated cells were protected from external damage compared with DAP-treated cells. and wanted to identify the genes involved in the effective cryopreservation of SCK-cryopreserved hiPS cells using DNA microarray analysis.
hiPS cells are commonly cryopreserved as cell colonies by vitrification [11,21]. After thawing, the cells are cultured as clots on a feeder layer without making a single cell suspension, but often do not survive this process [34]. Hence, we hypothesized that hiPS cells underwent apoptosis post-revival. In order to address this using DNA microarray data, we first checked the term ‘apoptosis’ in GO-BP, and compared the gene profile of SCK-cryopreserved hiPS cells with those of non-frozen or DAP-cryopreserved hiPS cells with respect to differences in gene expression. We extracted the genes that exhibited a difference in expression of more than 1.5-fold and found 15 genes; seven genes were anti-apoptotic in nature, while the remaining eight genes had an apoptotic function. Hence, both anti-apoptotic and apoptotic genes were competitively expressed in SCK-cryopreserved hiPS cells.
In contrast, we extracted only four anti-apoptotic genes that exhibited a difference in expression of more than 1.5-fold in DAP-cryopreserved hiPS cells versus non-frozen hiPS cells, while the remaining nine genes strongly facilitated apoptosis. Among the apoptotic genes extracted from SCK- and DAP-cryopreserved hiPS cells compared with non-frozen hiPS cells, BCL2A1, BID, PRIKCA, STK17B and MLH1 were commonly expressed, though they fell into in different categories from apoptosis in DAVID analysis. In the hsa04210 apoptosis KEGG pathway, SCK-cryopreserved hiPS cells exhibited three genes, BIRC3(LAPXIP), BID and CASP6, with expression differences higher than those of non-frozen hiPS cells. According to the KEGG pathway, BID and CASP6 enhance apoptosis, whereas BIRC3 suppresses CASP3, 7 and 9, and is a pro-survival gene in itself. This suggests that SCK-cryopreserved hiPS cells would exhibit significantly improved survival after thawing. This speculation needs to be inspected through further experimentation (Table 3a, b; Fig. 2d, Fig. S1).
When we compared the genes between SCK- and DAP-cryopreserved hiPS cells that exhibited a difference in expression of more than 1.5-fold, only one was selected in SCK-cryopreserved hiPS cells. Hence, a less stringent condition (i.e., change of more than 1.4-fold) was applied, allowing four genes (BIRC3, FADD, SOCS3, and BMF) to be extracted. While there were no effective gene categories extracted in DAVID, however, a strong gene network including BIRC3, FADD, and SOCS3 was formed by physical interaction, co-expression, pathways, and genetic interaction in GeneMANIA. Thus, our data indicated that the anti-apoptotic function of SCK-cryopreserved hiPS cells may be stronger than the apoptotic function.
The three keywords, ‘cell adhesion,’ ‘cell proliferation,’ and ‘stem cell,’ extracted by GO-BP are important factors that play a role in stem cell culture after their revival. We extracted the genes of SCK-cryopreserved hiPS cells in these keywords using a less stringent condition, (genes exhibiting a difference in expression of more than 1.4-fold), as we predicted that the differences between the expression of these genes would not be large. In case of the keyword ‘cell adhesion,’ 11 genes were extracted, of which four fell under the category of ‘cell adhesion’ when analyzed by DAVID. Furthermore, six of the genes exhibited a physical interaction in GeneMANIA. Hence, we concluded that the SCK-cryopreserved hiPS cells demonstrated strong adhesive properties after thawing.
In the case of the keyword, ‘cell proliferation,’ 19 genes were extracted. Out these, 15 were involved in a wide gene network, although their gene expression was not as strong in GeneMANIA. For the keyword ‘stem cell,’ we first used the keyword ‘stem cell maintenance.’ However, as only two genes were selected using this term, we changed the selecting condition to ‘stem cell’. We extracted four genes for this keyword, with two categories in BP-GO and two KEGG pathways found in DAVID analysis. However, no gene network was detected in GeneMANIA regarding the keyword ‘stem cell’. With respect to the gene expression associated with these keywords in SCK- and DAP-cryopreserved hiPS cells, a severe decline in expression was observed in the DAP-cryopreserved hiPS cells, while low to moderate gene expression was observed in the SCK-cryopreserved hiPS cells. Furthermore, these differences in the gene expression profiles could be associated with attachment of the hiPS cells to culture dishes after thawing, cell growth, and maintenance of pluripotency of the stem cells.
Further studies are required to investigate the expression of the individual genes that were reported in this study. Additionally, as only the 253G1 strain of hiPS cells was used in this study, it is important to investigate other hiPS cell strains as well to confirm the reproducibility of this study.
The final goal of this study was to determine the effective genes and their interactions in SCK-cryopreserved hiPS cells. We found that SCK-cryopreserved hiPS cells showed a group of anti-apoptotic genes and other groups related to the keywords ‘cell proliferation,’ ‘cell adhesion,’ and ‘stem cell,’ however, how these genes interact is still unknown. Further studies should research the effects of these genes on cryopreservation through Western blotting, immunochemical staining, reverse transcript PCR, or gene editing methods.
Recently, the development of chemical ice-inhibition molecules, including cryoprotectant, antifreeze protein, synthetic polymer, nanomaterial, and hydrogel, and their applications in regenerative devices and cryopreservation, has progressed. Additionally, advanced engineering strategies, including trehalose delivery, cell encapsulation, and bioinspired structure design for ice inhibition, are also amazingly developed [[41], [42], [43]]. Through the combination of SCK and these novel products or advanced engineering techniques, we expect to improve cryopreservation methods.
In conclusion, the DNA microarray analysis of SCK-cryopreserved hiPS cells demonstrated that apoptotic genes BID and CASP6, enhanced apoptosis, whereas BIRC3, an anti-apoptotic gene, suppressed CASP3, 7 and 9 in the apoptosis KEGG pathway. Owing to anti-apoptotic function of BIRC3 as well as genes involved in cell adhesion, cell proliferation, and multipotency, SCK-cryopreserved hiPS cells are likely to exhibit survival and easy culturing after thawing. Thus, SCK is likely superior to DAP for stem cell storage and maintenance. Our results showed that SCK is suitable for the efficient preservation of stem cells that can be used clinically and in basic research for regenerative medicine. While more genetic analysis is needed, we suggest SCK as a superior cryopreservation agent to DAP and more appropriate for clinical use and future investigations.
Data availability
Data will be made available on request.
Author disclosure statement
K.M. and S.-H.H. are cofounders of Bioverde Inc.; A.O. is an employee of Bioverde, Inc.
Declaration of competing interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Acknowledgements
The authors would like to acknowledge Mr. Kawakatsu, T (Kurabo Inc.) for his technical expertise. Funding: This study was supported in part by a Grant-in-Aid, KAKENHI (25242050, 20H04532), for Scientific Research from the Japan Society for the Promotion of Science.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101172.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Hochedlinger K., Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/DEV.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/J.STEM.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Baker M. iPS cells: potent stuff. Nat. Methods. 2010;7:17–19. doi: 10.1038/nmeth.f.281. [DOI] [PubMed] [Google Scholar]
- 4.Hunt C.J. Cryopreservation of human stem cells for clinical application: a review. Transfus. Med. Hemotherapy. 2011;38:107–123. doi: 10.1159/000326623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li T., Mai Q., Gao J., Zhou C. Cryopreservation of human embryonic stem cells with a new bulk vitrification method. Biol. Reprod. 2010;82:848–853. doi: 10.1095/BIOLREPROD.109.080713. [DOI] [PubMed] [Google Scholar]
- 6.Matsumura K., Hayashi F., Nagashima T., Hyon S.H. Long-term cryopreservation of human mesenchymal stem cells using carboxylated poly-l-lysine without the addition of proteins or dimethyl sulfoxide. J. Biomater. Sci. Polym. Ed. 2013;24:1484–1497. doi: 10.1080/09205063.2013.771318. [DOI] [PubMed] [Google Scholar]
- 7.Rall W.F., Fahy G.M. Ice-free cryopreservation of mouse embryos at −196 °C by vitrification. Nature. 1985;313:573–575. doi: 10.1038/313573a0. 1985 3136003. [DOI] [PubMed] [Google Scholar]
- 8.Kuleshova L.L., Gouk S.S., Hutmacher D.W. Vitrification as a prospect for cryopreservation of tissue-engineered constructs. Biomaterials. 2007;28:1585–1596. doi: 10.1016/J.BIOMATERIALS.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi A., Maehara M., Uchikura A., Matsunari H., Matsumura K., Hyon S.H., Sato M., Nagashima H. Development of an efficient vitrification method for chondrocyte sheets for clinical application. Regen. Ther. 2020;14:215–221. doi: 10.1016/J.RETH.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maehara M., Sato M., Watanabe M., Matsunari H., Kokubo M., Kanai T., Sato M., Matsumura K., Hyon S.-H., Yokoyama M., Mochida J., Nagashima H. Development of a novel vitrification method for chondrocyte sheets. BMC Biotechnol. 2013;13:58. doi: 10.1186/1472-6750-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumura K., Bae J.Y., Kim H.H., Hyon S.H. Effective vitrification of human induced pluripotent stem cells using carboxylated ε-poly-l-lysine. Cryobiology. 2011;63:76–83. doi: 10.1016/j.cryobiol.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Ota A., Matsumura K., Lee J.J., Sumi S., Hyon S.H. Stemcell keepTM is effective for cryopreservation of human embryonic stem cells by vitrification. Cell Transplant. 2017;26:773–787. doi: 10.3727/096368916X692654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumura K., Hyon S.H. Polyampholytes as low toxic efficient cryoprotective agents with antifreeze protein properties. Biomaterials. 2009;30:4842–4849. doi: 10.1016/j.biomaterials.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 14.Matsumura K., Bae J.Y., Hyon S.H. Polyampholytes as cryoprotective agents for mammalian cell cryopreservation. Cell Transplant. 2010;19:691–699. doi: 10.3727/096368910X508780. [DOI] [PubMed] [Google Scholar]
- 15.Matsumura K., Hatakeyama S., Naka T., Ueda H., Rajan R., Tanaka D., Hyon S.-H. Molecular design of polyampholytes for vitrification-induced preservation of three-dimensional cell constructs without using liquid nitrogen. Biomacromolecules. 2020;21:3017–3025. doi: 10.1021/ACS.BIOMAC.0C00293. [DOI] [PubMed] [Google Scholar]
- 16.Vorontsov D.A., Sazaki G., Hyon S.H., Matsumura K., Furukawa Y. Antifreeze effect of carboxylated ε-poly-l-lysine on the growth kinetics of ice crystals. J. Phys. Chem. B. 2014;118:10240–10249. doi: 10.1021/jp507697q. [DOI] [PubMed] [Google Scholar]
- 17.Matsumura K., Hayashi F., Nagashima T., Rajan R., Hyon S.-H. Molecular mechanisms of cell cryopreservation with polyampholytes studied by solid-state NMR. Commun. Mater. 2021;21(2):1–12. doi: 10.1038/s43246-021-00118-1. 2021. [DOI] [Google Scholar]
- 18.Adler S., Pellizzer C., Paparella M., Hartung T., Bremer S. The effects of solvents on embryonic stem cell differentiation. Toxicol. Vitro. 2006;20:265–271. doi: 10.1016/J.TIV.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 19.Chetty S., Pagliuca F.W., Honore C., Kweudjeu A., Rezania A., Melton D.A. A simple tool to improve pluripotent stem cell differentiation. Nat. Methods. 2013;10:553–556. doi: 10.1038/nmeth.2442. 2013 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagata N. Survival of mouse morulae and blastocysts derived from in vitro fertilization after ultrarapid freezing. Exp. Anim. 1993;42:229–231. [PubMed] [Google Scholar]
- 21.Fujioka T., Yasuchika K., Nakamura Y., Nakatsuji N., Suemori H. A simple and efficient cryopreservation method for primate embryonic stem cells. Int. J. Dev. Biol. 2004;48:1149–1154. doi: 10.1387/IJDB.041852TF. [DOI] [PubMed] [Google Scholar]
- 22.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. 2007 4487151. [DOI] [PubMed] [Google Scholar]
- 23.McMahon A.P., Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-R. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K., Okita K., Nakagawa M., Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2007;212(2):3081–3089. doi: 10.1038/nprot.2007.418. 2007. [DOI] [PubMed] [Google Scholar]
- 25.Tan P.K., Downey T.J., Spitznagel E.L., Jr., Xu P., Fu D., Dimitrov D.S., Lempicki R.A., Raaka B.M., Cam M.C. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res. 2003;31:5676–5684. doi: 10.1093/nar/gkg763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy D.J., Smyth G.K. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics. 2009;25:765–771. doi: 10.1093/BIOINFORMATICS/BTP053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/NAR/GKN923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;41(4):44–57. doi: 10.1038/nprot.2008.211. 2009. [DOI] [PubMed] [Google Scholar]
- 29.Gene Ontology Consortium Going forward 2015 the gene Ontology consortium list of authors of the gene Ontology consortium is provided in the appendix. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Expansion of the gene Ontology knowledgebase and resources 2017 the gene Ontology consortium list of authors of the gene Ontology consortium is provided in the appendix. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., Maitland A., Mostafavi S., Montojo J., Shao Q., Wright G., Bader G.D., Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/NAR/GKQ537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franz M., Rodriguez H., Lopes C., Zuberi K., Montojo J., Bader G.D., Morris Q. GeneMANIA update 2018. Nucleic Acids Res. 2018;46:W60–W64. doi: 10.1093/NAR/GKY311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh Y.-H., Wu Q., Chew J.-L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K.-Y., Sung K.W., Lee C.W.H., Zhao X.-D., Chiu K.-P., Lipovich L., Kuznetsov V.A., Robson P., Stanton L.W., Wei C.-L., Ruan Y., Lim B., Ng H.-H. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. 2006 384. [DOI] [PubMed] [Google Scholar]
- 34.Ichikawa H., Nakata N., Abo Y., Shirasawa S., Yokoyama T., Yoshie S., Yue F., Tomotsune D., Sasaki K. Gene pathway analysis of the mechanism by which the Rho-associated kinase inhibitor Y-27632 inhibits apoptosis in isolated thawed human embryonic stem cells. Cryobiology. 2011;64:12–22. doi: 10.1016/j.cryobiol.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz P.H., Brick D.J., Nethercott H.E., Stover A.E. Traditional human embryonic stem cell culture. Methods Mol. Biol. 2011;767:107–123. doi: 10.1007/978-1-61779-201-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergström R., Ström S., Holm F., Feki A., Hovatta O. Xeno-free culture of human pluripotent stem cells. Methods Mol. Biol. 2011;767:125–136. doi: 10.1007/978-1-61779-201-4_9. [DOI] [PubMed] [Google Scholar]
- 37.Kuleshova L., Gianaroli L., Magli C., Ferraretti A., Trounson A. Birth following vitrification of a small number of human oocytes: case Report. Hum. Reprod. 1999;14:3077–3079. doi: 10.1093/HUMREP/14.12.3077. [DOI] [PubMed] [Google Scholar]
- 38.Li Y., chun Tan J., song Li L. Comparison of three methods for cryopreservation of human embryonic stem cells. Fertil. Steril. 2010;93:999–1005. doi: 10.1016/J.FERTNSTERT.2008.10.052. [DOI] [PubMed] [Google Scholar]
- 39.Lee J.Y., Lee J.E., Kim D.K., Yoon T.K., Chung H.M., Lee D.R. High concentration of synthetic serum, stepwise equilibration and slow cooling as an efficient technique for large-scale cryopreservation of human embryonic stem cells. Fertil. Steril. 2010;93:976–985. doi: 10.1016/J.FERTNSTERT.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 40.Tan F.C.K., Kong H.L., Sok S.G., Magalhães R., Poonepalli A., Hande M.P., Dawe G.S., Kuleshova L.L. Optimization of cryopreservation of stem cells cultured as neurospheres: comparison between vitrification, slow-cooling and raped cooling “freezing” protocols. Cryo-Letters. 2007;28:445–460. [PubMed] [Google Scholar]
- 41.Chang T., Zhao G. Ice inhibition for cryopreservation: materials, strategies, and challenges. Adv. Sci. 2021;8 doi: 10.1002/ADVS.202002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmadian E., Eftekhari A., Dizaj S.M., Sharifi S., Mokhtarpour M., Nasibova A.N., Khalilov R., Samiei M. The effect of hyaluronic acid hydrogels on dental pulp stem cells behavior. Int. J. Biol. Macromol. 2019;140:245–254. doi: 10.1016/J.IJBIOMAC.2019.08.119. [DOI] [PubMed] [Google Scholar]
- 43.Khezri K., Maleki Dizaj S., Rahbar Saadat Y., Sharifi S., Shahi S., Ahmadian E., Eftekhari A., Dalir Abdolahinia E., Lotfipour F. Osteogenic differentiation of mesenchymal stem cells via curcumin-containing nanoscaffolds. Stem Cell. Int. 2021 doi: 10.1155/2021/1520052. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.