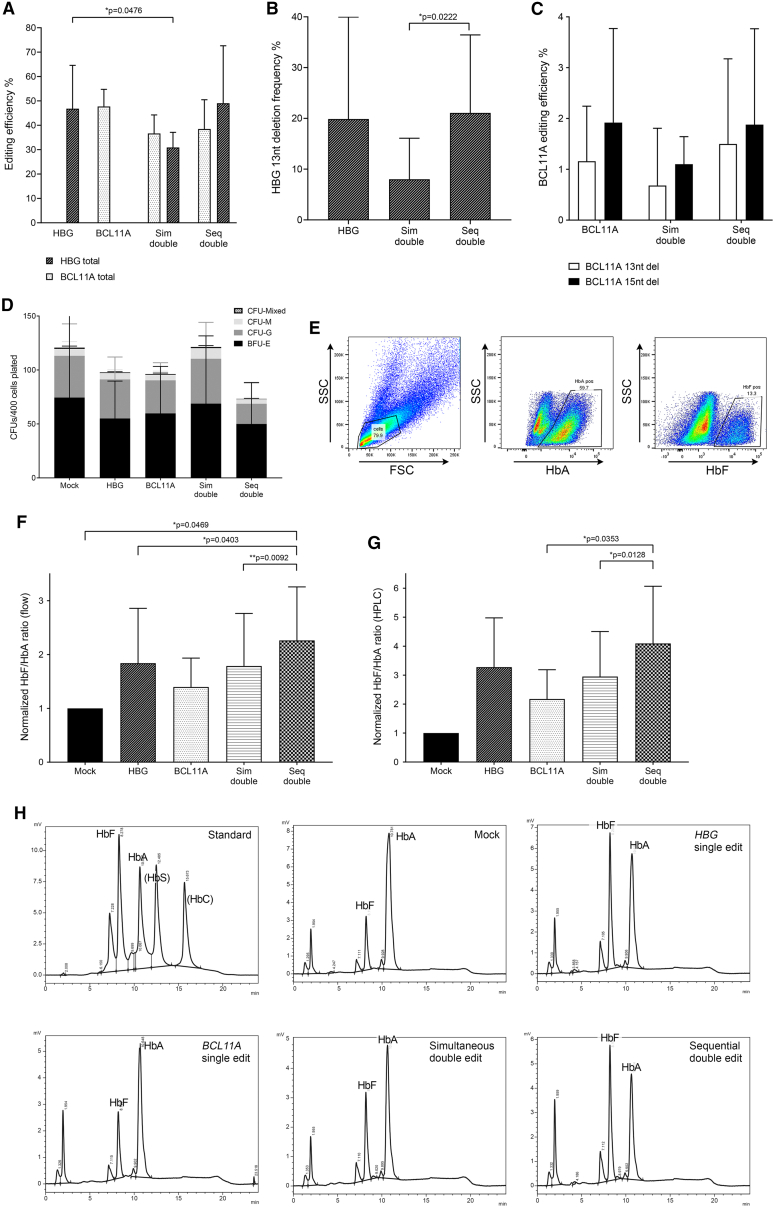
Figure 2.
Analysis of bulk CD34+ cells in vitro following single, dual, or mock editing
(A) Total HBG-113 and total BCL11A-ee editing efficiency in each reaction (n = 5) (mean ± SD). (B) Frequency of HBG-113 13 nucleotide deletion (n = 5) (mean ± SD). (C) Frequency of BCL11A-ee 13 and 15 nucleotide deletions (n = 5) (mean ± SD). (D) Colony-forming potential of CD34+ cells from each reaction (n = 5) (mean ± SD). (E) Sample flow cytometric plot demonstrating gating strategy to allow assessment of HbA and of HbF-positive cells within bulk cells grown in differentiation media. (F) Ratio of HbF-positive cells to HbA-positive cells in each reaction by flow cytometry, normalized to mock (n = 5) (mean ± SD). (G) Ratio of HbF percentage to HbA percentage in each reaction by HPLC, normalized to mock (n = 4) (mean ± SD). (H) Examples of HPLC traces from each reaction within a single experiment, as well as the HPLC standard demonstrating elution time of HbF and HbA. BCL11A, BCL11A-ee single-edited reactions; HBG, HBG-113 single-edited reactions; nt, nucleotide; Seq, sequential; Sim: simultaneous. Only significant differences (p < 0.05) are shown.