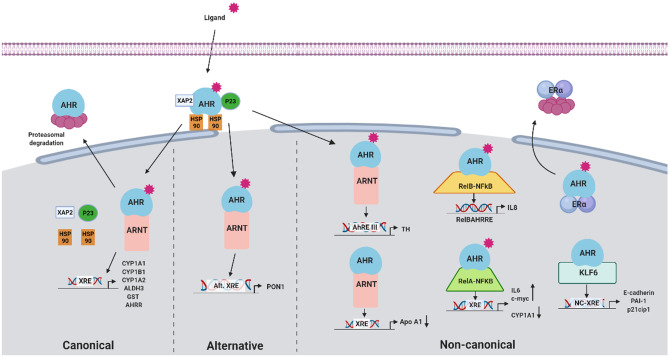
Fig. 1.
Representations of ligand-dependent AHR activation pathways. Inactive AHR is localized at the cytoplasm complexed to some chaperones and other elements (HSP90, XAP2, p23, c-Src). Upon ligand binding, conformational changes allow AHR to translocate to the nucleus, where it dissociates from its chaperone complex. Depending on the ligand, AHR can follow either ARNT-dependent or ARNT-independent pathways. In the ARNT-dependent pathways, the AHR-ligand complex dimerizes with its binding partner ARNT. The ligand-AHR-ARNT complex will activate or repress the expression of different genes depending on the type of ligand. Canonical pathway (left side). AHR binding to Kyn, HAH (e.g., dioxin), or PAH (e.g., B[a]P) activates the canonical pathway; the dimer AHR-ARNT binds to xenobiotic response elements (XRE) and drives the expression of xenobiotic metabolizing enzymes such as CYP1A1, the prototypical target gene of AHR. The AHR-ARNT complex promotes gene expression by recruiting several components of the transcriptional machinery and fulfilled this function; AHR activity ends by the dissociation of the complex from the DRE to be exported from the nucleus, where it enrolls in ubiquitin-mediated proteasomal degradation [26]. Moreover, a negative regulatory feedback mechanism is present, with the induction of AHR repressor (AHRR) gene, capable of competing with AHR for ARNT, originating a transcriptionally inactive heterodimer, thus repressing AHR transcriptional activity [13]. Alternative pathway (center). AHR binding to polyphenols such as resveratrol or quercetin activates the alternative pathway by binding to alternative xenobiotic response elements such as the antioxidant and anti-inflammatory paraoxonase 1 (PON-1). Other non-canonical pathways (right side) include alternative REs of alternative AHR-ARNT related transcriptional responses also observed in tyrosine hydroxylase [27], a precursor enzyme in the synthesis of dopamine and catecholamines, Bax, an apoptosis regulator gene [28], and in TGF-β [29]. Also, the AHR can bind to other non-ARNT binding partners, like the Kruppel-like factor 6 (KLF6) or the NF-kB subunits RelA or RelB. ALHD3, aldehyde dehydrogenase 3; AHR, aryl hydrocarbon receptor; AHRE, aryl hydrocarbon response element; AHRR, AHR repressor; ARNT, aryl hydrocarbon receptor nuclear translocator; B[a]P, Benzo[a]pyrene; CYP1A1, cytochrome P450, family 1, subfamily A, polypeptide 1; CYP1A2, cytochrome P450, family 1, subfamily A, polypeptide 2; CYP1B1, cytochrome P450, family 1, subfamily B, polypeptide 1; GST, glutathione S-transferase; HAH, halogenated aromatic hydrocarbon; HSP90, heat shock protein 90; KLF6, Kruppel-like factor 6; Kyn, kynurenine; NC-XRE, non-consensus xenobiotic responsive element; p21cip1, cyclin-dependent kinase inhibitor 1; p23, HSP90-associated co-chaperone; PAI-1, plasminogen activator inhibitor 1; PAH, polycyclic aromatic hydrocarbon; PON1, paraoxonase 1; RelA, nuclear factor NF-kappa-B P65 subunit; RelB, RELB proto-oncogene, NF-kB subunit; TH, tyrosine hydroxylase; XAP2, hepatitis B virus X-associated protein 2 or aryl hydrocarbon receptor-interacting protein (AIP); XRE, xenobiotic responsive element